Figure 6.
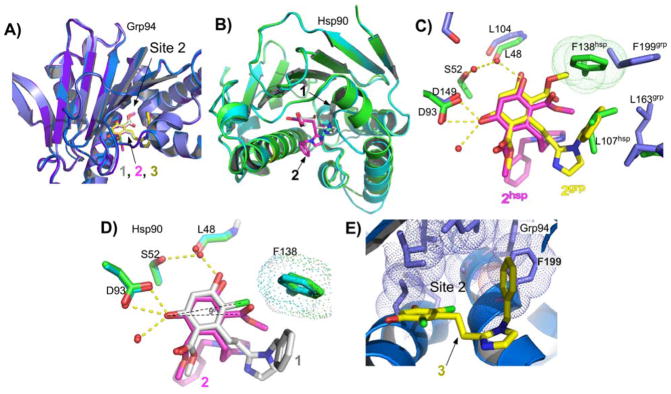
Binding of ligands 2 and 3 in Grp94 and Hsp90. A) Superposition of Grp94N:1, Grp94N:2, and Grp94N:3 showing similar poses for the ligand. B) The binding pose of 2 in Hsp90 is distorted. Overview alignment of Hsp90:1 (green) and Hsp90:2 (cyan) crystal structures showing the good superposition of the two protein structures except for the region between residues 103 and 111. The benzyl imidazole groups from 1 (grey) and 2 (magenta) are found in different poses in the two structures. C) Superposition of Grp94N:2 and Hsp90N:2 showing the different poses for the bound ligand and potential clashes between the Grp94 pose and the Hsp90 pocket. D) The resorcinol scaffolds of 1 (gray) and 2 (magenta) make similar hydrogen bonds in the Hsp90 ATP binding pocket but 2 is rotationally displaced by about 9 degrees in order to prevent clashes between the larger methyl ester group and Phe138, which blocks Site 2 in Hsp90. E) Phe199 in Grp94 exposes Site 2 in Grp94N:3 despite having the smaller chloro moieties on the resorcinol ring. The residues of Site 2 are shown as sticks with dot surfaces.