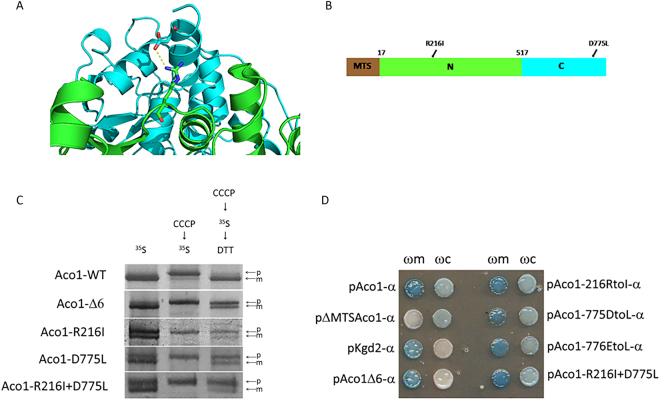
Figure 3.
Post-translational import of mutant Aco1 and N/C domains. (A) The predicted salt bridge between Aco1 N and C terminal domains, based on a model generated by the I-TASSER software. N terminal domains, green; C terminal domain, light blue; Predicted salt bridge, yellow. (B) Schematic illustration of aconitase predicted salt-bridge mutations. MTS, brown; N terminal domain with the R216I mutation, green; C terminal domain with the D775L mutation, light blue. (C) Single mutations in the predicted salt bridge affect import into mitochondria. Wild-type yeast cultures harboring the indicated plasmids were induced in galactose medium and labelled with [35S] methionine in presence of CCCP which blocks import (left lanes). Following labelling, aliquots of these cultures were chased with excess cold methionine/cysteine in the presence of DTT for an additional 10 min (right lanes). Cell lysates were than immunoprecipitated with aconitase antiserum and analyzed by SDS-PAGE followed by autoradiography. Precursor, p; Mature, m. (D) Alpha complementation of the predicted salt bridge mutants. Yeast cultures co-expressing cytosolic ω (ωc) or mitochondrial ω (ωm) together with various α-fused proteins were grown on galactose medium containing X-gal. Blue colonies indicate α fragments that are associated with the ω fragments. Controls: pKgd2- α (dihydrolipoyl transsuccinylase 2) as a mitochondrial marker; pΔMTSAco1- α (Aconitase lacking mitochondrial targeting signal) as cytosolic marker.