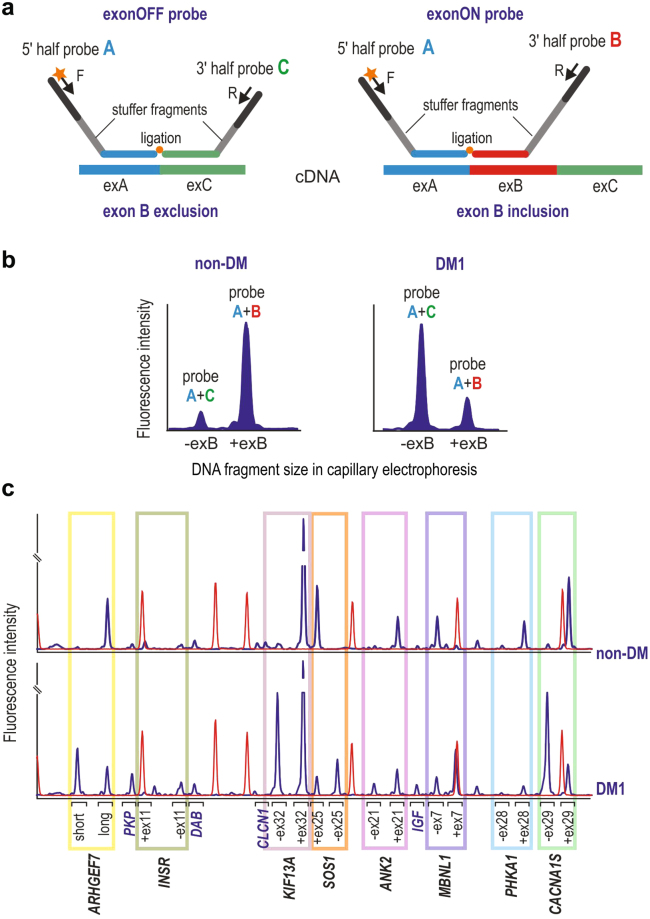
Figure 2.
MLPA Assays Designed. (a) Schematic representation of the ex-OFF probe (left panel) and the ex-ON probe (right panel) specifically hybridized to the cDNA of a transcript, respectively, without and with the alternative exon B. Exons A and C are constitutive exons flanking the alternative exon B. Each of the exon-specific probes is composed of a 5′ half-probe and a 3′ half-probe (5′ half-probe is shared by both ex-OFF and ex-ON probes). Each half-probe is composed of a target-specific sequence (indicated by a color corresponding to targeted exon), stuffer sequence (gray), and sequence specific to either F (labeled) or R universal primers. Note that stuffer sequence in 3′ half-probe is different in size in the ex-OFF and ex-ON probes allowing differentiating the signal of probes in capillary electrophoresis. (b) Comparison of ssMLPA signals (electropherogram peaks) of the hypothetical exon-specific pair of probes (shown in panel a) in non-DM and DM1 samples (note, different ratio of signals of ex-ON and ex-OFF probes in compared samples). The ratio of signals from both peaks was used to calculate the PSI value for each sample. (c) Representative ssMLPA results (electropherograms) of non-DM and DM1 samples. Blue peaks represent signals of MLPA probes used for indicated genes. The color rectangles indicate the pairs of exon-specific probes. The signals outside the rectangles represent control probes. Red peaks denote GS Liz600 DNA size standard.