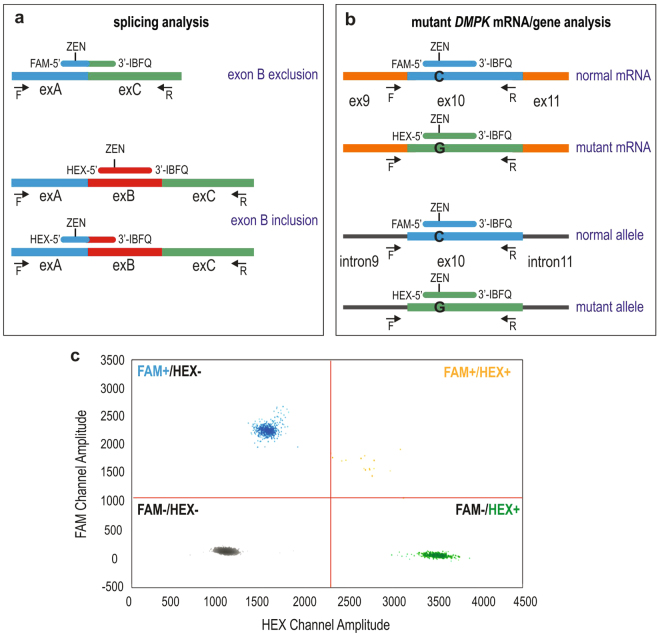
Figure 3.
ddPCR Probes Used. (a) An alternative exon exclusion probe (FAM-labeled) and two alternative exon inclusion probes (HEX-labeled) used in alternative splicing assays; the probes have ZEN-Iowa Black as the dual-quencher. An alternative exon (exB) is indicated in red, and it’s flanking exons (exA and exC) in blue and green. Location of primers (F and R) is shown by arrows and ZEN probes are displayed in their binding exons. (b) Dual-quencher probes used in DMPK copy number quantification in cDNA (upper part) and gDNA (lower part). The rs527221 SNP in exon 10 of DMPK was used to distinguish normal-size and mutant alleles. Position of primers (F and R) is shown by arrows and probes are displayed in their binding sites. (c) Representative 2-dimensional scatter plot displaying droplet populations in separate clusters depending on their fluorescence amplitude following ddPCR for CACNA1S exon 29 (DM2 sample). FAM-positive droplets (FAM+/HEX−), HEX-positive (FAM−/HEX+), double-positive (FAM+/HEX+), and negative droplets (FAM−/HEX−) are shown.