Figure 3.
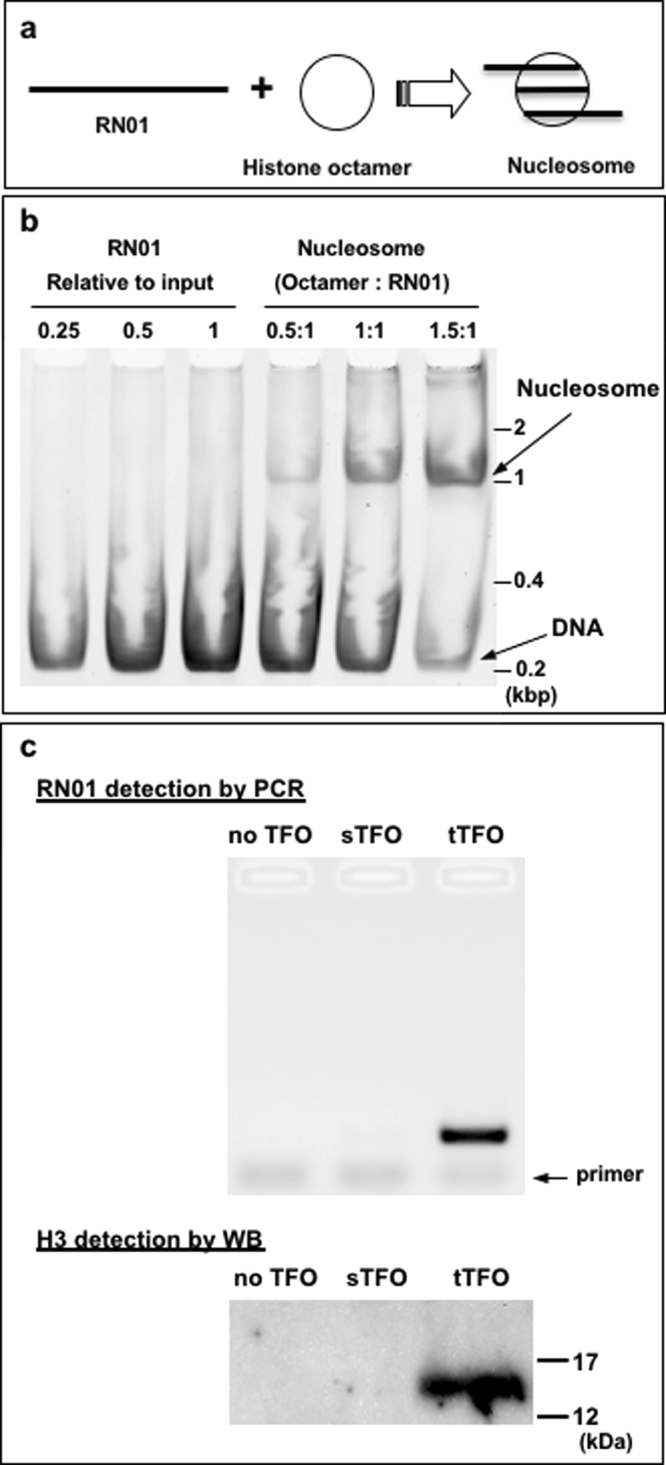
Nucleosome capture depends upon the presence of tTFO: (a) Schematic view of reconstitution of mono-nucleosome. (b) A double-stranded (209 bp) DNA (RN01) is used for reconstituting mono-nucleosomes in vitro. RN01 is incubated with the component of the histone core (i.e., human histones H2A, H2B, H3, and H4). Gel shift assay is carried out to assess mono-nucleosome reconstitution: octamer / DNA ratio ranging from 0.5 to 1.5. The results show that >50% of DNA is incorporated into mono nucleosomes at octamer/DNA ratio equal to one or above. (c) Cross-linked RN01 nucleosomes are incubated with buffer (no TFO), scrambled TFO (sTFO: a mixture of Scr-1 and Scr-2) the sequences unable to form a triplex with the TFT cassette, or target TFO (tTFO: a mixture of TFO-1 and TFO-3) complementary to the TFT cassette for triplex formation. These samples are subsequently incubated with streptavidin-conjugated beads. Beads are washed and bound material is eluted, followed by heat to reverse the cross-links. A small fraction (0.001%) of each elution product is amplified by PCR for DNA detection, followed by quantification using Image Lab software (Bio-Rad). PCR quantification was performed as detailed in Fig. 6d. We performed three independent experiments that all led to results similar to those shown in the present figure. For western blotting (WB), 80% of the elution products are used for histone H3 detection. Full-length blots are presented in Supplementary Fig. S8.
