Fig. 3.
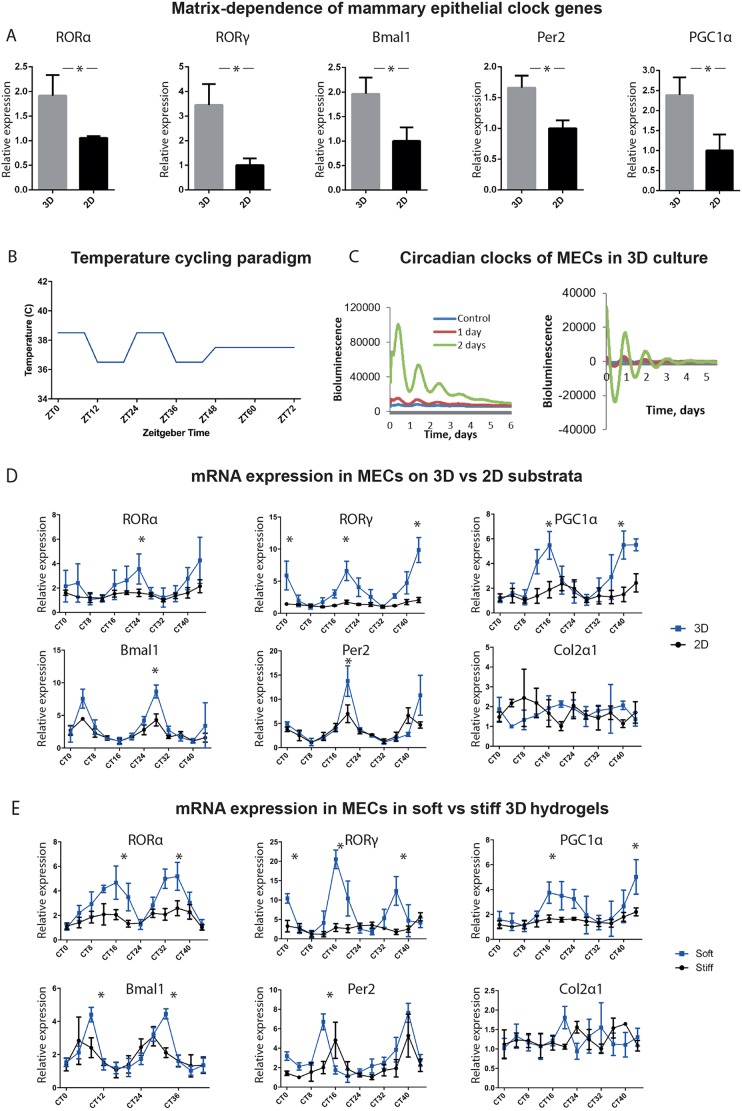
Expression of clock genes is mechano-sensitive in MECs. (A) Expression of genes encoding RORα, RORγ, Bmal1, Per2 and PGC1α is matrix-dependent, with higher levels of expression in MECs cultured in 3D vs 2D culture. n=3 biological replicates, each with 3 mice, pooled to form 3 cultures per condition, with a further 3 technical replicates for each gene. (Student's t-test, P<0.05 and P<0.01 for RORα; mean±s.e.m.). (B) Temperature cycling paradigm consists of 48 h in a temperature-controlled incubator, which cycles between 36.5°C and 38.5°C every 12 h, followed by 24 h at 37°C. For RT-qPCR, sampling begins thereafter (named as circadian time 0, CT0), every 4 hours until CT44. (C) MECs cultured in 3D show robust circadian oscillation that increases as the cells are in temperature cycles for longer times prior to recording. Bioluminescence traces presented as raw (left) and normalised (right). n=3 biological replicates, each with 4 mice, pooled to form 2 cultures per condition. (D) Time-course of clock and clock-controlled genes is significantly different in 3D vs 2D cultures. There is higher circadian mRNA expression in 3D for RORα, RORγ, PGC1α, Bmal1 and Per2 than in cells on 2D substrata. Note that there is no difference in the circadian expression of genes known not to be under circadian control in MECs, such as collagen2α1. n=3 biological replicates, each with 6 mice, pooled to form 36 cultures per condition. (Student's t-test, P<0.05, mean±s.e.m.). (E) Time-course of clock and clock-controlled genes is significantly different in cells in 3D soft versus stiff alginate gels. There is higher circadian expression in soft 3D for RORα, RORγ, PGC1α, Bmal1 and Per2, than in cells in the stiff gels. n=3 biological replicates, each with 16 mice, pooled to form 36 cultures per condition. (Student's t-test, P<0.05, mean±s.e.m.).