Abstract
Leaf color mutants in higher plants are ideal materials for investigating the structure and function of photosynthetic system. In this study, we identified a cucumber vyl (virescent-yellow leaf) mutant in the mutant library, which exhibited reduced pigment contents and delayed chloroplast development process. F2 and BC1 populations were constructed from the cross between vyl mutant and cucumber inbred line ‘Hazerd’ to identify that the vyl trait is controlled by a simply recessive gene designated as CsVYL. The CsVYL gene was mapped to a 3.8 cM interval on chromosome 4 using these 80 F2 individuals and BSA (bulked segregation analysis) approach. Fine genetic map was conducted with 1542 F2 plants and narrowed down the vyl locus to an 86.3 kb genomic region, which contains a total of 11 genes. Sequence alignment between the wild type (WT) and vyl only identified one single nucleotide mutation (C→T) in the first exon of gene Csa4G637110, which encodes a DnaJ-like zinc finger protein. Gene Expression analysis confirmed the differences in transcription level of Csa4G637110 between wild type and mutant plants. Map-based cloning of the CsVYL gene could accelerate the study of chloroplast development and chlorophyll synthesis of cucumber.
Keywords: Cucumis sativus L., leaf color mutant, virescent, chloroplast function, DnaJ proteins
Introduction
Leaf is a vital organ for plant photosynthesis that occurs mainly in chloroplasts. Chloroplast, a semi-autonomous organelle exits only in plant cells, can synthesize various metabolites that are essential for plant growth and development, and maintenance of green leaf color (Moreira et al., 2000). Leaf color mutants are formed due to silencing or inactivation of genes controlling chlorophyll biosynthesis and chloroplast development, which directly or indirectly affects chlorophyll synthesis and degradation, ultimately leading to leaf color variation (Motohashi et al., 2003). Therefore, leaf color mutants are the ideal materials to study the mechanism of plant photosynthesis, chloroplast ultrastructure, chlorophyll biosynthetic pathway, expression and regulation of related genes (Stern et al., 2004). So far, leaf color mutants have been obtained in rice (Zhang et al., 2006), wheat (Falbel et al., 1996), maize (Lonosky et al., 2004), soybean (Zhang et al., 2011), cotton (Karaca et al., 2004), carrot (Nothnagel and Straka, 2003) and cucumber (Gao et al., 2016). Numerous studies have been reported in relative genetic, physiological and molecular mechanism research.
Virescent, which shows light yellow cotyledons or true leaf and turns green gradually during leaf development, is a special and important mutation type for understanding the mechanism of chloroplast formation and most of virescent mutant are thermo- or light-sensitive (Archer and Bonnett, 1987; Yoo et al., 2009). A few virescent mutants have been reported in Arabidopsis (Koussevitzky et al., 2007), rice (Sugimoto et al., 2007), maize (Hopkins and Elfman, 1984) and other plants. Several genes have been positional cloned in these plants. In rice, Dong et al. (2013) used a temperature-insensitive and developmental stage-dependent virescent yellow leaf (vyl) mutant to clone the vyl gene related to plastidic caseinolytic protease. Xing et al. (2014) reported a virescent yellow-like (vyl) maize mutant that exhibited young yellow leaf phenotype but gradually recovered, and identified a gene Chr.9_ClpP5 as the candidate gene, which is one of the proteolytic subunits of the chloroplast Clp protease complex. In cucumber, Miao et al. (2016) used a virescent leaf mutant 9110Gt showing light yellow cotyledons and first true leaf to detect a single recessive gene (v-1) controlled the virescent leaf trait, and identified a most likely candidate gene for the v-1 by high-resolution genetic mapping.
DnaJ proteins are important molecular chaperones in the cell and play significant roles in protein folding, assembly, localization, transportation and denatured protein renaturation or degradation (Hartl, 1996; Walsh et al., 2004). A 41 kDa heat shock protein DnaJ was first found in Escherichia coli (Georgopoulos et al., 1980), and subsequent studies have shown that DnaJ proteins are ubiquitously found in humans, animals, plants and fungi (Zhu et al., 1993; Terada et al., 1997; Miernyk, 2001). Canonical DnaJ proteins consist of a J-domain, a glycine/phenylalanine (G/F) domain, a zinc finger domain (included four repeats of the CxxCxGxG sequence) and a C-terminal domain (Walsh et al., 2004). Different from the DnaJ proteins, DnaJ-like zinc finger proteins lack of characteristic J-domain and G/F domain of typical DnaJ proteins structure, and only have one zinc finger domain in the C-terminal (CxxCxGxG sequence repeat in 2–4 times) (Lu et al., 2006). Previous studies have shown that most of the DnaJ-like zinc finger proteins have important regulatory functions for plastid development, photosynthesis, chloroplast movement, plant resistance and defense response (Shimada et al., 2007; Fristedt et al., 2014; Muñoz-Nortes et al., 2017).
Cucumber, Cucumis sativus L. (2n = 2x = 14) is an important vegetable crop worldwide. Although many studies of leaf color mutants have been reported in cucumber (Pierce and Wehner, 1990), little work has been performed on map-based cloning of mutant gene or explaining its regulation mechanism. In this study, a novel light-sensitive virescent-yellow leaf cucumber mutant, vyl, was identified in an ethyl methanesulfonate (EMS)-mutagenized Changchunmici (‘CCMC’, north China type) cucumber population. The mutant exhibited virescent yellow leaf at emergence with decreased chlorophyll accumulation and impaired chloroplast structure. Yellow leaf would turn green during development or under low light condition. The present study describes the results on phenotypic, physiological characterization and genetic of the cucumber vyl mutant and fine mapping of the candidate gene, CsVYL, which encodes a DnaJ-like zinc finger protein. These findings may facilitate the understanding of chloroplast development and chlorophyll synthesis in cucumber.
Materials and Methods
Plant Materials and Phenotypic Data Collection
The virescent yellow-leaf (vyl) mutant was isolated from an M2 family derived from an EMS-mutagenized cucumber ‘Changchunmici’ (CCMC, wild type, WT, a common inbred line with green leaf) population. Mutagenized plants of this second generation were self-pollinated for two generations to make the mutated gene homozygous, and then an F2 and BC1 populations were produced with the vyl (female) and green leaf cucumber inbred line ‘Hazerd’ (male) as the parents. The M4 generation, along with the F1, F2, and BC1 generations, were subsequently utilized to elucidate the inheritance pattern of vyl. A total of 80 individuals from F2 population were used for BSA analysis and then another 400 and 1065 individuals from F2 population were planted to fine map the vyl gene. All plants were grown in the greenhouse at Jiangpu Cucumber Research Station of Nanjing Agricultural University, Nanjing, China.
The leaf color of each plant was determined by visual inspection at 3–5 days after emergence. Chi Square Goodness of fit test was performed on phenotypic data to verify deviations from the expected 3:1 segregation in the F2 population or 1:1 segregation in the BC1 population.
Measurement of Pigment Contents and Photosynthetic Parameters
Leaves of vyl mutant and wild type plants at four different leaf positions (leaf 1, 2, 3, and 4) were collected when the plants grow to the stage with 6–7 true leaves and used to determine chlorophyll content according to the method described by Arnon (1949) with some modifications. Chlorophyll a and chlorophyll b were calculated according to the formula:
Total carotenoid content of the leaf was measured as described by Soto-Zamora et al. (2005). The absorbance was determined in the spectrophotometer at 470 nm.
In addition, photosynthetic parameters including net photosynthetic rate (Pn), stomatal conductance (Gs), and intercellular CO2 concentration (Ci) of vyl mutant and wild type plants were measured in the field from 9:00 to 12:00 on the sunny day using the US-made LI-6400 portable photosynthesis measurement system. Chlorophyll fluorescence was measured using a MINI-PAM (Walz, Effeltrich, Germany). The maximum quantum efficiency of Photosystem II (PSII), Fv/Fm = (Fm–Fo)/Fm (where Fo is the minimal and Fm the maximal fluorescence yield in dark-adapted leaves), was measured at the beginning of the day from leaves that were dark-adapted for 30 min in dark leaf clips DLC-8 (Walz, Effeltrich, Germany). The measurement was performed on the first leaf to the fourth leaf leaves on each plant in the same way. There individuals were measured on each repeat at four leaf positions and then the average was taken.
Different Light Intensity Treatment
We found that vyl might be a light-sensitive mutant, because its leaf color changed as light intensity changed. To verify the correlation between vyl leaf color and light intensity, WT and vyl mutant were planted under three different levels of light intensity conditions, through the use of shading. Three light intensity treatments included full light (no shading, about 65000 LUX at the noon), 68% of full light, and 3% of full light. The corresponding light intensity was measured by the digital illuminometer Peakmeterm-S6612. The phenotype of vyl under different light intensity treatments was identified as well as its chlorophyll contents.
Transmission Electron Microscopy
Leaf samples of wild type and vyl mutant were prepared for TEM from the 1st, 2nd, 3rd, and 4th leaf with the blade when the 4th leaf expanded fully. Leaf sections were vacuumized and fixed in 2.5% glutaraldehyde in a phosphate buffer at 4°C for 4 h. The samples were dehydrated in a graded ethanol series, and critical-point drying was done using liquid CO2 in a critical-point drier Bal Tec CPD 030 and 15 nm gold coated on aluminum stubs in a Sputte Coater Bal-Tec SCD 005. The samples were visualized with a Hitachi S-3500N scanning electron microscope.
DNA Extraction and BSA
Young leaf samples of each individual were collected and frozen in liquid nitrogen and maintained at -80°C for further use in experiments. Genomic DNAs were extracted from leaves by the modified cetyltrimethylammonium bromide (CTAB) method of Murray and Thompson (1980). For the bulked segregant analysis (BSA), 10 individuals with virescent yellow color leaf and 10 with green color were randomly selected from the 80 plants of F2 population, and two DNA bulks were made by bulked equal quantities of DNA of the 10 individuals.
Molecular Marker Development and Mapping Strategy
In initial mapping phase, 450 published SSR (simple sequence repeat) markers evenly distributed on 7 chromosomes were selected randomly from the SSR library of ‘9930’ (Ren et al., 2009) and ‘Gy14’ (Cavagnaro et al., 2010) to screen for polymorphisms between the two parents and two pools. Then polymorphism markers were used to identify the genotypes of each individual of 80 plants from the F2 mapping population and construct an initial genetic map. When no recombination events was observed between the flanking markers, the size of the mapping population was expanded to 480 plants for further testing (including the initial 80 plants). Additional SSR markers in the preliminary mapping region were developed by screening the relative sequences of ‘9930’ genome, which were available from the Cucurbit Genomics Database1, using the SSR Hunter 1.3 program.
A larger F2 population (n = 1545, including initial 480 plants) were applied to fine map the CsVYL locus. Two parents, vyl mutant and ‘Hazard,’ were re-sequenced by the Illumina Hi-Seq 4000 platform to develop more available molecular markers. Short reads obtained from two parents were aligned against the cucumber genome sequence (the ‘9930’ reference genome) to obtain the consensus sequence using BWA software (Huang et al., 2009; Langmead and Salzberg, 2012). Reads were aligned to ‘9930’ consensus sequence reads to call SNPs with SAM tools software (Li and Durbin, 2009). The correlated sequences of two mapping parents in the initial interval were used to develop Indel and CAPS molecular markers. Insertions and deletions (>3 bp) were screened using the SAM Tool program in this interval to design Indel markers by Primer 5.0. In addition, the SNPs were converted into CAPS (cleaved amplified polymorphic sequences) markers using SNP2CAPS program. Finally, polymorphic markers between two parents were further screened from the newly developed markers and used to identify recombinants in the 1545 plants of the F2 population for fine mapping the CsVYL locus. Primers of molecular makers used in fine mapping of CsVYL are listed in Supplementary Table 1.
The linkage map was constructed using Join Map 4.0 with a LOD threshold score of 3.0. The PCR reaction and gel electrophoresis was conducted as described in Li et al. (2011).
Gene Annotation and Candidate Gene Identification
Candidate genes in target region were predicted using the online program FGENESH2 and the Cucumber Genome Database3 (Huang et al., 2009) and their annotations were obtained with the BLASTP tool from NCBI4. For the candidate gene identified, the DNA, cDNA and protein sequences alignments were performed using the software DNAMAN to detect variations between vy1 and WT. And in order to verify the reliability of candidate gene, the DNA sequence of candidate gene were sequencing among 72 natural cucumber lines. Protein sequences of CsVYL and its homologous were downloaded from NCBI database. Primers used for cloning of candidate gene sequence are listed in Supplementary Table 2.
Quantitative Reverse-Transcription PCR (qPCR) Analysis
Total RNA was extracted from the first (yellow stage) and fifth leaves (green stage) of CCMC (WT) and vyl (representing yellow and green stages) with RNAprep Plant Mini Kit (Tiangen, Beijing, China) following the manufacturer’s instructions. First-strand cDNA was synthesized using Prime Script 1st Strand cDNA Synthesis Kit (TaKaRa, Kyoto, Japan). To analyze gene expression, quantitative real-time PCR (qPCR) was carried out using the Real Master Mix Kit (SYBR Green, Tiangen). qRT-PCR and PCR amplification was quantified according to the manufacturer’s protocol. All experiments were performed with three biological replicates. The 2-ΔΔCT method was used to analyze the relative mRNA expression level of vyl, and the values represented the n-fold difference relative to the gene expression of WT at the yellow stage. Primers used for qPCR are listed in Supplementary Table 3.
Results
The vyl Is a Development Dependent and Light Sensitive Mutant
A mutant with virescent yellow-leaf (vyl) was identified in an M2 generation from EMS-mutagenized ‘CCMC’ population which exhibited virescent yellow leaf at emergence and then gradually recovered (Figures 1A,C). Different from leaf color mutant reported previously (Miao et al., 2016), the mutant studied in this work exhibited virescent yellow leaf only in true leaves whereas the leaf vein remains green (Figure 1B). At maturity, the vyl mutant could normally blossom, bear fruits and produce seeds (Figure 1C).
FIGURE 1.
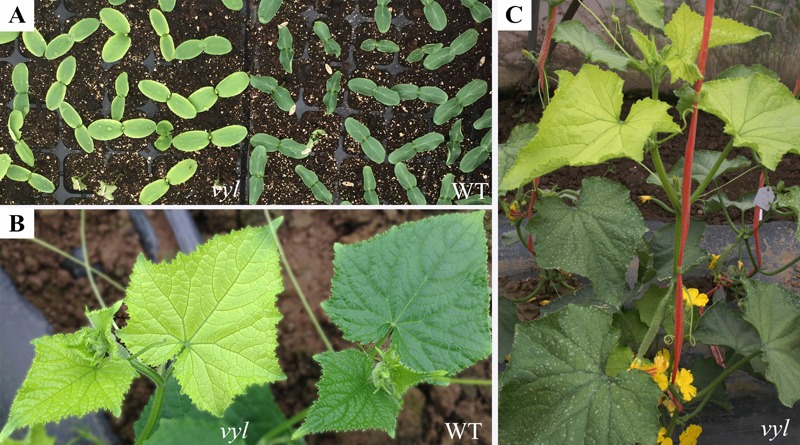
Phenotypic analysis of the vyl mutant. (A) Cotyledons color of WT and vyl; (B) Detailed observations of leaf color of WT and vyl grown in field; (C) Leaf color of vyl at plant adult stage.
To investigate whether the photosynthetic apparatus was affected in vyl mutants, we compared the content of some key photosynthetic pigments between vyl and WT plants (Table 1). The contents of chloroplyll a, chloroplyll b and carotenoid were basically consistent in WT at different leaf positions, whereas the contents of the photosynthetic pigments in vyl mutant were significantly decreased compared to those in WT at early leaf positions (the first to the third leaf from the top). When the leaf grows, leaf color turned green gradually and photosynthetic pigments increased to reach the level in WT after the fourth leaf. To verify whether vyl is a light-sensitive mutant, WT and vyl were planted under different light intensity conditions and the change of leaf color and Chl contents of vyl was examined. As shown in Figure 2A, under 100% full light condition, compared with the WT plants, the vyl mutant exhibited extreme chlorosis and its Chl contents were lower than those in WT. As the vyl mutant grew under reduced light intensity condition, leaf color of vyl showed a greening trend, and Chl contents also increased (Figures 2B,C). Notably, under extreme low light condition (about 3% full light), the vyl mutant showed the same leaf color phenotype and Chl contents as WT under the same conditions (Figure 2C). These results indicated that virescent-yellow leaf in this mutant is a light sensitive trait.
Table 1.
Comparison of pigment content between vyl and the wild type at different leaf position.
| Leaf | Materials | Chloroplyll | Chloroplyll | Carotenoid |
|---|---|---|---|---|
| position | a (mg/g) | b (mg/g) | (mg/g) | |
| L1 | Wild type | 1.1 ± 0.12 | 0.46 ± 0.10 | 0.44 ± 0.07 |
| vyl | 0.40 ± 0.08** | 0.14 ± 0.03** | 0.20 ± 0.05** | |
| L2 | Wild type | 1.14 ± 0.09 | 0.46 ± 0.09 | 0.44 ± 0.08 |
| vyl | 0.54 ± 0.05** | 0.20 ± 0.05** | 0.24 ± 0.07** | |
| L3 | Wild type | 1.14 ± 0.18 | 0.44 ± 0.06 | 0.42 ± 0.04 |
| vyl | 0.66 ± 0.09** | 0.24 ± 0.04** | 0.26 ± 0.04** | |
| L4 | Wild type | 1.14 ± 0.12 | 0.44 ± 0.04 | 0.42 ± 0.05 |
| vyl | 0.92 ± 0.09 | 0.36 ± 0.02** | 0.34 ± 0.03 |
∗∗Means significant differences at 0.01 level. L1, L2, L3, L4 indicate different leaf position, for example, L1 means the first leaf below the plant top.
FIGURE 2.
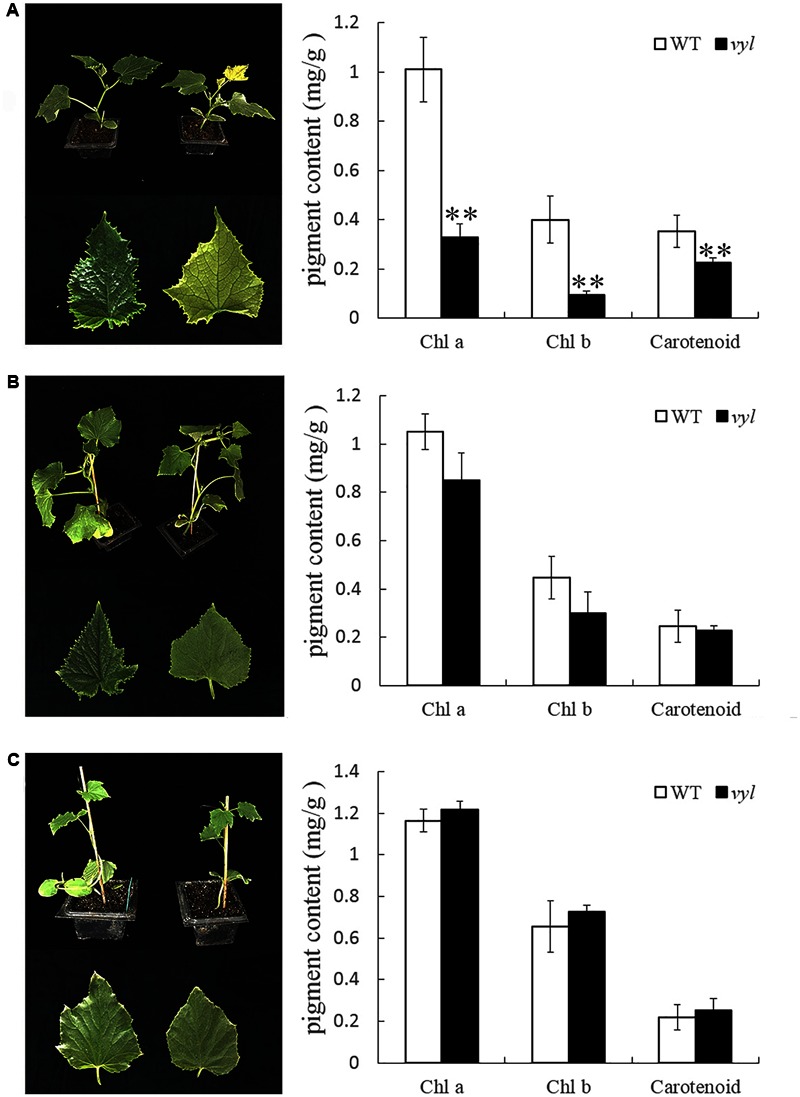
The leaf color phenotype and pigment contents of vyl and WT under different light intensity treatments. L1 of vyl and WT planted under 100% (A), 68% (B) and 3% (C) of full light conditions was used for leaf color observation and Chl measurement. Values are the mean ± SD, ∗∗P < 0.01.
The vyl Has Defective Chloroplast Development and Reduced Photosynthetic Capacity at Early Leaf Stage
Mesophyll cells of the first to the fourth leaf (from the plant top) from vyl mutant and WT were examined by TEM. The impaired chloroplast development with various degrees in the mutant was observed, especially in the first to the third leaf, and recovered to the similar level as WT eventually, after the fourth leaf (Figures 3A–D), whereas the WT had a morphologically well-structured thylakoid system at each leaf position (Figures 3E–H). Notably, membrane system was outspread and the thylakoids were not stacked into grana at the first leaf in vyl. As photosynthetic assimilation products, starch grains were hardly observed at the first leaf in vyl (Figure 3A), which was clearly visible in WT (Figure 3E). This result also indicated the difference of photosynthetic function between vyl mutant and WT. At the second leaf in the mutant (Figure 3B), stacked grana and starch grain were observed. As the leaf turned green at the third leaf (Figure 3C), more stacked grana and starch grain were observed in vyl. At the fourth leaf in vyl (Figure 3D), there was more starch grain and the stacked grana exhibiting well developed lamellar structures, similar to that in WT (Figure 3H).
FIGURE 3.
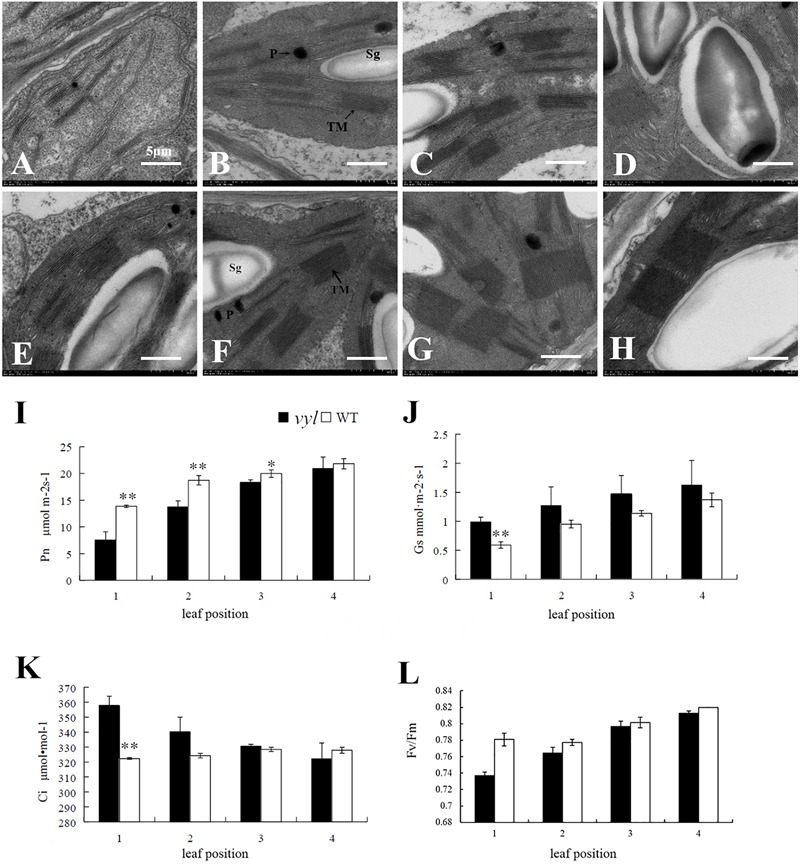
Chloroplast structure and photosynthesis parameters of vyl and WT at different leaf position. Electron microscopy of chloroplasts ultrastructures in L1-L4 of vyl (A–D) and WT (E–H). TM, thylakoid membrane; Sg, starch grain; P, plastoglobulus. Scale bar = 500 nm. (I) Net photosynthetic rate (Pn); (J) Stomatal conductance (Gs); (K) Intercellular CO2 concentration (Ci); (L) Photosystem II (Fv/Fm); Values are the mean ± SD, and ∗(P < 0.05) and ∗∗(P < 0.01) indicate significant difference between the treatments.
Several key photosynthetic parameters were compared at different leaf positions in vyl mutant and WT plants (Figure 3). Young leaves of vyl mutant showed significantly lower Pn than WT plants (Figure 3I). Because both ‘stomatal’ and ‘non-stomatal’ components can contribute to the decrease of Pn (Quick et al., 1992), and the value of sub-stomatal CO2 concentration (Ci) can be used to distinguish the two components (Farquar and Sharkey, 1982). Higher Gs and Ci suggest that the decrease of Pn in vyl mutant young leaves was caused by non-stomatal component (Figures 3J,K). As leaf matured, vyl mutant and WT plants displayed similar photosynthesis, indicating the recovery of photosynthesis (Figure 3). Similar trend was also observed in the maximum quantum efficiency of PSII (Fv/Fm). Fv/Fm was lower in the young leaf of vyl mutant than in WT plants, suggests that the photosystem of vyl mutant was undeveloped compared to the contemporaneous WT plants (Figure 3L).
Genetic Mapping of CsVYL Gene Using BSA Method
To determine the inheritance patterns of virescent yellow-leaf trait, the vyl was crossed with ‘Hazerd’ (green leaf) to generate F2 and BC1 populations. The F1 individuals have green leaf, and among the 80 plants of F2 population, there are 58 green leaf individuals and 22 vyl individuals, corresponding with the 3:1 ratio of Mendel’s law of segregation (χ2 = 0.27, p = 0.61). For the 100 plants derived from backcross with the vyl parent, 53 had green leaf and 47 were vyl leaf, conforming to the 1:1 ratio of Mendel’s law of segregation (χ2 = 0.36, p = 0.55). The segregation ratios obtained above supported that a single recessive gene, CsVYL, controls the virescent yellow leaf trait.
Genetic mapping of the CsVYL gene was performed using bulked segregation analysis approach. First, among the 450 SSR markers evenly distributed on seven cucumber chromosomes, 84 polymorphic SSR markers (18.7%) between the parents, vyl and ‘Hazerd,’ were screened. Then using two DNA sample pools from vyl mutant and green leaf individuals of the F2 population as templates, the 84 markers were used to determine linkage relationship between the markers and vyl phenotype. Four SSR markers (UW084200, UW042029, SSR05515 and UW084372) were observed to be linked with the vyl phenotype. These four SSR markers were used to genotype 80 F2 individuals and CsVYL locus was located within an approximately 3.8 cM interval between UW084200 and SSR05515, co-segregating with UW042029 on distal end of the long arm of chromosome 4 (Figure 4A). Since no recombination events was observed between the flanking markers, the size of mapping population was expanded to 480 plants for further testing (including the initial 80 plants). To narrow down the target region, the genomic sequence of ‘9930’ in the target interval was used to develop new SSR markers. Totally 184 pairs of SSR makers were developed in this region and 10 polymorphic markers (5.4%) were detected. All polymorphic markers, two flanking markers, UW084200 and SSR05515 and one co-segregating marker (UW042029) were used to construct a local genetic map of the CsVYL locus using 480 F2 individuals. Two closest markers, SSR60 and SSR113, flanked the CsVYL locus at a 0.8 cM interval (∼467 kb), and a co-segregating marker, SSR63 was found (Figure 4B).
FIGURE 4.
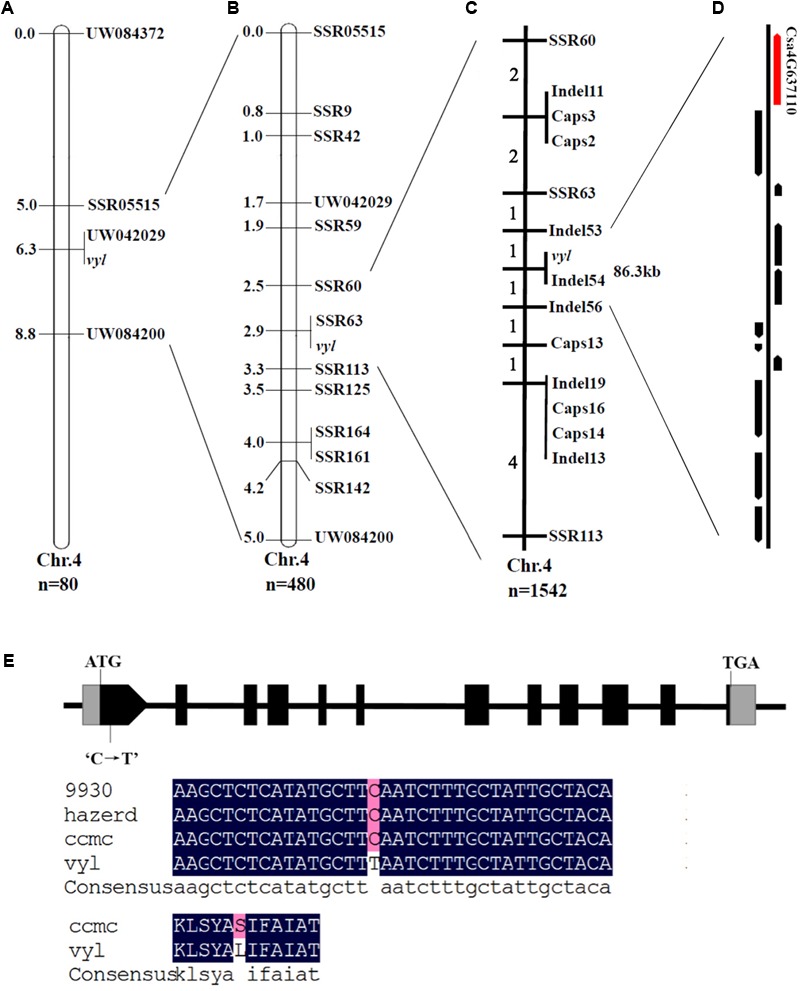
Positional cloning and sequence alignment of the CsVYL gene. (A) Preliminary mapping with BSA based on 80 plants from the F2 population placed CsVYL locus on chromosome 4 at a 3.8 cM region between UW804200 and SSR05515. Numbers on chromosome left are genetic distance (cM); (B) Further genetic position analysis with 480 F2 individuals delimited the CsVYL locus at a 0.8 cM region; (C) Fine mapping of the CsVYL locus with 1542 individuals. The CsVYL gene was narrowed down to an 86.3 kb genetic region. Numbers on chromosome left are recombinants of each interval; (D) 11 annotated genes were predicted in the 86.3 kb region. Csa4G637110 is shown as the red arrow. (E) Gene and protein sequence alignment of CsVYL gene in 9930, Hazerd, ccmc and vyl Gray boxes, black boxes and lines between them indicate 5′ and 3′UTR, exons and introns respectively.
Fine Mapping Using Genome Sequence Data
Since additional effort of exploring markers in the region flanked by SSR60 and SSR113 couldn’t develop new markers that were closer to the target gene, we conducted high-depth re-sequencing of two mapping parents, vyl mutant and ‘Hazerd,’ to develop Indel and SNP markers (The Illumina sequence raw data have been submitted to NCBI SRA database, accession number SRP134028, and re-sequencing data is listed in Supplementary Table 4). Indels that varied > 3 bp in length were selected to develop Indel markers. Totally 74 Indel markers were developed, among which, 6 markers that showed obvious polymorphism between two parents were used to fine map the CsVYL locus. In addition, 35 CAPS makers were developed by screening SNP between the target region and 5 of them were polymorphic between two parents. Using flanking markers, SSR60 and SSR113, 13 recombinant plants were selected from 1542 plants from the F2 population. Eleven newly developed markers, two flanking markers and one co-segregating marker were subsequently used to map the CsVYL locus with the 13 recombinant plants. Finally, fine genetic mapping placed the CsVYL locus in the genomic region flanked by markers Indel53 and Indel56, which was approximately 86.3 kb in cucumber reference genome (Figure 4C).
Candidate Gene Identification
In the 86.3 kb region, 11 genes were annotated using the online program FGENESH and ‘9930’ genome sequence data (Figure 4D). The information and predicted functions of the 11 genes were presented in Table 2. Coding sequences of these genes were sequenced and aligned between WT and vyl mutant, and a single nucleotide mutation occurred in gene Csa4G637110 was found, whereas no differences were found in other 10 genes. Besides, the single nucleotide mutation of the candidate gene existed only in the vyl mutant, but not in other 72 natural populations (Supplementary Figure 1). Annotation of Csa4G637110 revealed that there were 31 exons and 11 introns in this gene. The genomic sequence of this gene from WT and vy1 was cloned, and the full length of this gene is 8149 bp in ‘9930’ reference genome. Sequence alignment between WT and vy1 indicated that a single nucleotide mutation (C→T) occurred in the first exon of the Csa4G637110 gene, which resulted in an amino acid change of glutamic to lysine acid (Figure 4E and Supplementary Figure 2).
Table 2.
Predicted genes in the 86.3 kb region.
| ORF No. | Gene ID | Position in 9930 genome | Gene annotation |
|---|---|---|---|
| 1 | Csa4G637110 | 20695774..20703922 | DnaJ-like zinc finger protein |
| 2 | Csa4G637120 | 20705767..20713243 | Bifunctional methylthioribulose-1-phosphate dehydratase |
| 3 | Csa4G637130 | 20716210..20717384 | VQ motif-containing protein |
| 4 | Csa4G637140 | 20730318..20734988 | Dynamin-related protein |
| 5 | Csa4G637150 | 20735565..20739818 | Proton pump-interactor |
| 6 | Csa4G637160 | 20744740..20746545 | Xylosyltransferase |
| 7 | Csa4G637170 | 20746608..20747443 | Acetylglucosaminyltransferase |
| 8 | Csa4G637180 | 20748636..20750371 | Pentatricopeptide repeat-containing protein |
| 9 | Csa4G637680 | 20751673..20758425 | SPA1-related protein |
| 10 | Csa4G637690 | 20766468..20776209 | Uncharacterized protein |
| 11 | Csa4G637700 | 20780049..20785461 | Ubiquitin-conjugating enzyme |
ID number given by cmb (http://cmb.bnu.edu.cn/Cucumis_sativus_v20).
Protein sequences of the encoded protein and other 9 homologs were aligned. Detail alignment showed that all proteins have an incomplete zinc finger domain with two C-x(2)-C-x-G-x-G repeats and incomplete C-x(2)-C-x-G or C-x(2)-C repeats, different from the typical zinc finger domain of DnaJ protein which has four C-x(2)-C-x-G-x-G repeats (Supplementary Figure 3).
Expression Analysis of the Csa4G637110 and Other Related Genes
The expression level of Csa4G637110 gene was measured at different leaf color stages (yellow and green stages) from vyl mutant and WT to analyze the relationship between expression patterns of Csa4G637110 gene and the change of leaf color. According to qRT-PCR results, at the yellowing leaf stage of vyl mutant, the expression level of this gene in WT was significantly higher than that in vyl mutant leaves. Subsequently, the high expression level in WT decreased as the leaves aged and finally was equal to that in the vyl mutant (Figure 5).
FIGURE 5.
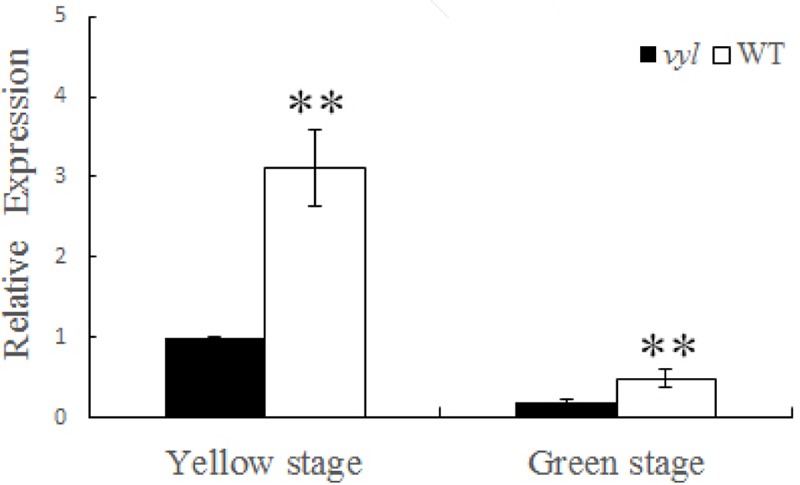
Relative expression of Cs4G637110 gene at different leaf color stages (yellow and green stage) of WT and vyl mutant by qRT-PCR. Data were displayed using the CsActin gene as an internal control with three biological and three technical replicates. Values are the mean ± SD. ∗∗P < 0.01.
The expression of the transcription of 12 genes involved in Chl biosynthesis and photosynthesis were analyze at seeding stage. The result was showed in Figure 6. There genes involved in Chl biosynthesis were down-regulated in vyl, including HEMA1 (glutamyl-tRNA reductase), POR (NADPH-dependent protochlorophyllide oxidoreductase) and CAO (Chlorophyllide a oxygenase). In contrast, compared with the WT, the expression level of PPOX (protoporphyrinogen oxidase) was significantly increased in the vyl mutant. It is worthwhile to note that most of the expression levels of chloroplast genome genes were reduced in the vyl mutant, including psaA, psaB, psbA, and psbB (subunit genes of PS I and PS II core complex), rbcL (rubisco large subunit gene) and rpoB (plastid RNA polymerase gene). There was no obvious difference in the expression of HEMA2 and rpoA (plastid RNA polymerase alpha subunit). The down-regulated expression levels of these genes implied that the mutant gene might involve in the Chl biosynthesis and photosynthesis pathway.
FIGURE 6.
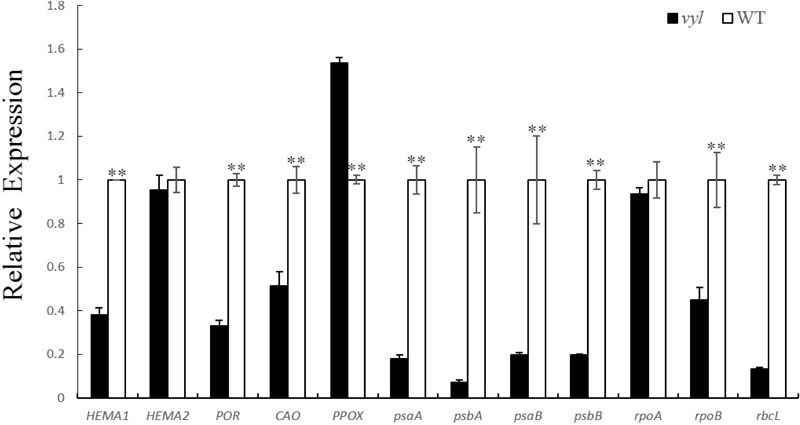
Expression evaluation of genes associated with Chl biosynthesis and photosynthesis through qRT-PCR. The L1 Leaves of vyl and WT were sampled. CsActin was used as the internal control. The transcript levels of all tested genes in wild type (WT) were set to 1.0. Values are the mean ± SD with three biological and three technical replicates. ∗∗P < 0.01.
Discussion
The main sources of leaf color mutants are natural mutagenesis or artificially induced mutagenesis, and most of leaf color mutants are controlled by recessive genes and occur at seedling stage (Pierce and Wehner, 1990). Some chlorotic types gradually turn green during the later leaf growth stage (Austin and Webber, 2005; Li et al., 2014), while some remained chlorosis in the whole development process which are usually lethal (Zhang et al., 2013). The virescent leaf color mutant used in this article was selected from M2 generation of ‘Changchunmici’ mutant library, which was caused by EMS mutagenesis. This virescent yellow leaf was regulated by a single recessive gene verified by genetic analysis. This vyl mutant showed yellow young leaf with obvious green leaf veins (Figure 1B), which is different with previous cucumber leaf color mutant v (Pierce and Wehner, 1990) and v-1 (Miao et al., 2016). Also, different from the whole-life leaf color mutant (Gao et al., 2016), the leaf color of this vyl mutant gradually turned green as the leaves age, and the plant can develop, blossom and yield fruit normally. The changes of leaf color in vyl plant from young leaf to mature leaf were accompanied by changes in chlorophyll content and photosynthetic capacity (Figure 3 and Table 1). Furthermore, we also verified that it was a light-sensitive mutation which showed virescent leaf only under strong light condition (Figure 2).
The mechanism of leaf chlorotic mutation is complicated and can generally be summarized into two possibilities. The first one is that the mutation of chlorophyll biosynthesis or degradation related gene directly leads to the change of pigment content in leaves, like OsCHLI and OsCHLD mutants (Zhang et al., 2006); the second possibility is that the mutation of chloroplast developmental genes directly leads to incomplete chloroplast development, therefore indirectly affects chlorophyll synthesis, like ASL2 (Lin et al., 2015), CSP41 (Mei et al., 2017) mutant gene in rice. In cucumber, the chlorophyll-deficient material, C528, has a golden leaf color throughout the whole life and chlorophyll content is lower in mutant than that in wild type. The corresponding gene, CsChlI, which encodes a CHLI subunit of cucumber Mg-chelatase, was cloned (Gao et al., 2016). Miao et al. (2016) used a virescent leaf color material (9110Gt) that shows incomplete chloroplast development, to clone a chloroplast development related gene, CsaCNGCs, which encodes a cyclic-nucleotide-gated ion channel protein. In this study, the chloroplast structure and chlorophyll content of the mutant were different from the wild type at yellowing stage. The grana stacks appeared sparse and irregular in shape, the thylakoid expanded largely, and assimilated products were less than in wild type (Figure 3A). The pigment of plant leaves was mainly located on the thylakoid of chloroplast. The chloroplast structure of the chlorotic mutant was damaged during chloroplast formation, which resulted in the abnormal development of thylakoid, eventually leading to decreased chlorophyll content compared to wild type (Motohashi et al., 2003). As the leaves were aging, the lamellar structure of grana stacks increased and the shape became regular. Chloroplast also gradually developed completely, the pigment levels were improved and leaves turned green similar to the wild type (Figures 3B–D). The delayed development of chloroplast is similar to that of 9110Gt (Miao et al., 2016), therefore, it can be speculated that the cause of yellowing young leaf is a result of damaged chloroplast structure.
For genetic mapping of CsVYL locus, chromosome walking combined with BSA and re-sequencing technology were adopted. In recent years, a number of phenotype-related genes have been cloned using the same strategy in cucumber (Li et al., 2013; Yang et al., 2014; Zhou et al., 2015). As a gene mapping method, it can quickly and accurately delineate target genes on an approximate position (Michelmore et al., 1991). Using two pools, 4 polymorphic SSR markers were screened from 450 SSR evenly distributed on 7 cucumber chromosomes. Then genotyping 80 plants of F2 population using the 4 markers allowed rapidly mapping the mutant gene in a 3.8 cM interval of chromosome 4. Next sequencing technology provides a powerful and cost-effective tool for exploring polymorphic markers during fine mapping (Liu et al., 2016). In this study, the parents (vyl and ‘Hazerd’) were re-sequenced to develop Indel and SNP markers in the target region, and finally the candidate gene was fine mapped in an 86.3 kb interval of chromosome 4, flanking by Indel53 and Indel56.
A total of 11 genes were detected in the 86.3 kb region. Through sequence alignment, a non-synonymous mutation leading to an amino acid sequence changes was found in Csa4G637110 coding region, suggesting that the mutation identified is responsible for the virescent-yellow leaf phenotype of vyl (Supplementary Figure 1). In addition, the function of this candidate gene was also verified by expression analysis. The transcription level of Csa4G637110 in vyl was significantly lower than that of WT during the young leaf stage. This high expression in WT reduced significantly when leaves aged. The expression pattern is similar to that of v-1 gene in cucumber (Miao et al., 2016), where a single-nucleotide mutation results in reduced expression of CsaCNGCs, and causes the occurrence of leaf color change. In addition, qPCR analysis results showed that most genes involved in Chl biosynthesis and photosynthesis were down-regulated in the vyl mutant at seeding stage (Figure 6). These findings indicated the important function of mutant gene in chloroplast development of cucumber at seeding stage. These findings suggested that Csa4G637110 is a plausible candidate of CsVYL. However, complementary experiments are needed to confirm its reliability.
The cucumber DnaJ-like zinc finger protein and its orthologs have a similar zinc finger domain containing the CxxCxGxGx repeats structure (Supplementary Figure 2), which is commonly found in the DnaJ protein. However, different from the typical DnaJ protein, DnaJ-like zinc finger proteins does not contain specific J Domain of DnaJ proteins and G/F domain structure, and Muñoz-Nortes et al. (2017) classified it as the type IV DnaJ protein. As a companion protein of HSP70 (heat shock protein 70), DnaJ protein and HSP70 are composed of the molecular chaperone that involved in many cellular processes, including protein folding, assembly, translocation and degradation of misfolded proteins (Georgopoulos, 1992; Hartl et al., 1994; Craig et al., 2006). Many biological processes require the participation of this molecular chaperon in Chloroplast, such as chloroplast development, photosynthesis, chloroplast protein transport (Vitha et al., 2003; Latijnhouwers et al., 2010; Kong et al., 2014). Recent studies have shown that DnaJ-like zinc finger proteins also have similar functions, and several mutants of these proteins resulted in different leaf color changes, for example, lqy1 and sco2-1 in Arabidopsis (Lu et al., 2011; Tanz et al., 2012), orange in cauliflower (Li et al., 2001), psa2 in maize (Fristedt et al., 2014). It can be speculated that the DnaJ-like zinc finger protein also has the same function in chloroplast development in cucumber. However, the regulatory mechanisms are still needed to be further researched.
Author Contributions
QL and JC conceived the research and designed the experiments. MS and JW performed the research. MS, QW, JW, and QL analyzed the data and wrote the manuscript. XQ participated in genome re-sequencing analysis. WF, XL, FC and KY were involved in phenotypic selection, DNA extraction, and mapping work. LZ, JL, and XY provided valuable experimental methods. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Martin Njogu (Department of Horticulture, College of Horticulture, Nanjing Agricultural University, Nanjing, China) for critical reading of the manuscript.
Funding. This research was partially supported by the National Natural Science Foundation of China (General Programs 31772318 and 31430075), the Fund for Independent Innovation of Agricultural Science and Technology of Jiangsu Province CX(17)3016, and the National Supporting Programs (2016YFD0100204-25) from the Ministry of Science and Technology of China.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00432/full#supplementary-material
References
- Archer E. K., Bonnett H. T. (1987). Characterization of a virescent chloroplast mutant of tobacco. Plant Physiol. 83 920–925. 10.1104/pp.83.4.920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon D. I. (1949). Copper enzymes in isolated chloroplasts. polyphenoloxidase in Beta vulgaris. Plant Physiol. 24 1–15. 10.1104/pp.24.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J. R., II, Webber A. N. (2005). Photosynthesis in Arabidopsis thaliana mutants with reduced chloroplast number. Photosynth. Res. 85 373–384. 10.1007/s11120-005-7708-x [DOI] [PubMed] [Google Scholar]
- Cavagnaro P. F., Senalik D. A., Yang L., Simon P. W., Harkins T. T., Kodira C. D., et al. (2010). Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genomics 11:569. 10.1186/1471-2164-11-569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E. A., Huang P., Aron R., Andrew A. (2006). The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 156 1–21. 10.1007/s10254-005-0001-0 [DOI] [PubMed] [Google Scholar]
- Dong H., Fei G. L., Wu C. Y., Wu F. Q., Sun Y. Y., Chen M. J., et al. (2013). A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol. 162 1867–1880. 10.1104/pp.113.217604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falbel T. G., Meehl J. B., Staehelin L. A. (1996). Severity of mutant phenotype in a series of chlorophyll-deficient wheat mutants depends on light intensity and the severity of the block in chlorophyll synthesis. Plant Physiol. 112 821–832. 10.1104/pp.112.2.821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquar G. D., Sharkey T. D. (1982). Stomatal conductance and photosynthesis. Annu. Rev. Plant Physiol. 33 317–345. 10.1146/annurev.pp.33.060182.001533 [DOI] [Google Scholar]
- Fristedt R., Williams-Carrier R., Merchant S. S., Barkan A. (2014). A thylakoid membrane protein harboring a DnaJ-type zinc finger domain is required for photosystem I accumulation in plants. J. Biol. Chem. 289 30657–30667. 10.1074/jbc.M114.587758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M. L., Hu L. L., Li Y. H., Weng Y. Q. (2016). The chlorophyll-deficient golden leaf mutation in cucumber is due to a single nucleotide substitution in CsChlI for magnesium chelatase I subunit. Theor. Appl. Genet. 129 1961–1973. 10.1007/s00122-016-2752-9 [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. (1992). The emergence of the chaperone machines. Trends Biochem. Sci. 17 295–299. 10.1016/0968-0004(92)90439-G [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P., Lundquist-Heil A., Yochem J., Feiss M. (1980). Identification of the E. coli DnaJ gene product. Mol. Gen. Genet. 178 583–588. 10.1007/BF00337864 [DOI] [PubMed] [Google Scholar]
- Hartl F. U. (1996). Molecular chaperones in cellular protein folding. Nature 381 571–579. 10.1038/381571a0 [DOI] [PubMed] [Google Scholar]
- Hartl F. U., Hlodan R., Langer T. (1994). Molecular chaperones in protein folding: the art of avoiding sticky situations. Trends Biochem. Sci. 19 20–25. 10.1016/0968-0004(94)90169-4 [DOI] [PubMed] [Google Scholar]
- Hopkins W. G., Elfman B. (1984). Temperature-induced chloroplast ribosome deficiency in virescent maize. J. Hered. 75 207–211. 10.1093/oxfordjournals.jhered.a109913 [DOI] [Google Scholar]
- Huang S., Li R., Zhang Z., Li L., Gu X., Fan W., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41 1275–1281. 10.1038/ng.475 [DOI] [PubMed] [Google Scholar]
- Karaca M., Saha S., Callahan F. E., Jenkins J. N., Read J. J., Percy R. G. (2004). Molecular and cytological characterization of a cytoplasmic-specific mutant in pima cotton (Gossypium barbadense L.). Euphytica 139 187–197. 10.1007/s10681-004-3032-3 [DOI] [Google Scholar]
- Kong F. Y., Deng Y. S., Zhou B., Wang G. D., Wang Y., Meng Q. W. (2014). A chloroplast-targeted DnaJ protein contributes to maintenance of photosystem II under chilling stress. J. Exp. Bot. 65 143–158. 10.1093/jxb/ert357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S., Stanne T. M., Peto C. A., Giap T., Sjogren L. L., Zhao Y., et al. (2007). An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 63 85–96. 10.1007/s11103-006-9074-2 [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/Nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers M., Xu X. M., Moller S. G. (2010). Arabidopsis stromal 70-kDa heat shock proteins are essential for chloroplast development. Planta 232 567–578. 10.1007/s00425-010-1192-z [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Paolillo D. J., Parthasarathy M. V., DiMuzio E. M., Garvin D. F. (2001). A novel gene mutation that confers abnormal patterns of beta-carotene accumulation in cauliflower (Brassica oleracea var. botrytis). Plant J. 26 59–67. 10.1046/j.1365-313x.2001.01008.x [DOI] [PubMed] [Google Scholar]
- Li Q., Zhu F. Y., Gao X., Sun Y., Li S., Tao Y., et al. (2014). Young Leaf Chlorosis 2 encodes the stroma-localized heme oxygenase 2 which is required for normal tetrapyrrole biosynthesis in rice. Planta 240 701–712. 10.1007/s00425-014-2116-0 [DOI] [PubMed] [Google Scholar]
- Li Y., Wen C., Weng Y. (2013). Fine mapping of the pleiotropic locus B for black spine and orange mature fruit color in cucumber identifies a 50 kb region containing a R2R3-MYB transcription factor. Theor. Appl. Genet. 126 2187–2196. 10.1007/s00122-013-2128-3 [DOI] [PubMed] [Google Scholar]
- Li Y., Yang L., Pathak M., Li D., He X., Weng Y. (2011). Fine genetic mapping of cp: a recessive gene for compact (dwarf) plant architecture in cucumber, Cucumis sativus L. Theor. Appl. Genet. 123 973–983. 10.1007/s00122-011-1640-6 [DOI] [PubMed] [Google Scholar]
- Lin D., Jiang Q., Zheng K., Chen S., Zhou H., Gong X., et al. (2015). Mutation of the rice ASL2 gene encoding plastid ribosomal protein L21 causes chloroplast developmental defects and seedling death. Plant Biol. 17 599–607. 10.1111/plb.12271 [DOI] [PubMed] [Google Scholar]
- Liu H., Jiao J. Q., Liang X. J., Liu J., Meng H. W., Chen S. X., et al. (2016). Map-based cloning, identification and characterization of the w gene controlling white immature fruit color in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 129 1247–1256. 10.1007/s00122-016-2700-8 [DOI] [PubMed] [Google Scholar]
- Lonosky P. M., Zhang X., Honavar V. G., Dobbs D. L., Fu A., Rodermel S. R. (2004). A proteomic analysis of maize chloroplast biogenesis. Plant Physiol. 134 560–574. 10.1104/pp.103.032003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Van Eck J., Zhou X., Lopez A. B., O’Halloran D. M., Cosman K. M., et al. (2006). The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. Plant Cell 18 3594–3605. 10.1105/tpc.106.046417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Hall D. A., Last R. L. (2011). A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23 1861–1875. 10.1105/tpc.111.085456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J. S., Li F. F., Liu X. R., Hu G. C., Fu Y. P., Liu W. Z. (2017). Newly identified CSP41b gene localized in chloroplasts affects leaf color in rice. Plant Sci. 256 39–45. 10.1016/j.plantsci.2016.12.005 [DOI] [PubMed] [Google Scholar]
- Miao H., Zhang S., Wang M., Wang Y., Weng Y., Gu X. (2016). Fine mapping of virescent leaf gene v-1 in cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 17:E1602. 10.3390/ijms17101602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R. W., Paran I., Kesseli R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88 9828–9832. 10.1073/pnas.88.21.9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk J. A. (2001). The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones 6 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D., Le Guyader H., Philippe H. (2000). The origin of red algae and the evolution of chloroplasts. Nature 405 69–72. 10.1038/35011054 [DOI] [PubMed] [Google Scholar]
- Motohashi R., Ito T., Kobayashi M., Taji T., Nagata N., Asami T., et al. (2003). Functional analysis of the 37 kDa inner envelope membrane polypeptide in chloroplast biogenesis using a Ds-tagged Arabidopsis pale-green mutant. Plant J. 34 719–731. 10.1046/j.1365-313X.2003.01763.x [DOI] [PubMed] [Google Scholar]
- Muñoz-Nortes T., Perez-Perez J. M., Ponce M. R., Candela H., Micol J. L. (2017). The ANGULATA7 gene encodes a DnaJ-like zinc finger-domain protein involved in chloroplast function and leaf development in Arabidopsis. Plant J. 89 870–884. 10.1111/tpj.13466 [DOI] [PubMed] [Google Scholar]
- Murray M. G., Thompson W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8 4321–4325. 10.1093/nar/8.19.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothnagel T., Straka P. (2003). Inheritance and mapping of a yellow leaf mutant of carrot (Daucus carota). Plant Breed. 122 339–342. 10.1046/j.1439-0523.2003.00884.x [DOI] [Google Scholar]
- Pierce L. K., Wehner T. C. (1990). Review of genes and linkage groups in cucumber. HortScience 25 605–615. [Google Scholar]
- Quick W. P., Chaves M. M., Wendler R., David M., Rodrigues M. L., Passaharinho J. A., et al. (1992). The effect of water-stress on photosynthetic carbon metabolism in 4 species grown under field conditions. Plant Cell Environ. 15 25–35. 10.1111/j.1365-3040.1992.tb01455.x [DOI] [Google Scholar]
- Ren Y., Zhang Z., Liu J., Staub J. E., Han Y., Cheng Z., et al. (2009). An integrated genetic and cytogenetic map of the cucumber genome. PLoS One 4:e5795. 10.1371/journal.pone.0005795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H., Mochizuki M., Ogura K., Froehlich J. E., Osteryoung K. W., Shirano Y., et al. (2007). Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. Plant Cell 19 3157–3169. 10.1105/tpc.107.051714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Zamora G., Yahia E. M., Brecht J. K., Gardea A. (2005). Effects of postharvest hot air treatments on the quality and antioxidant levels in tomato fruit. LWT Food Sci. Technol. 38 657–663. 10.1016/j.lwt.2004.08.005 [DOI] [Google Scholar]
- Stern D. B., Hanson M. R., Barkan A. (2004). Genetics and genomic of chloroplast biogenesis: maize as a model system. Trends Plants Sci. 9 293–301. 10.1016/j.tplants.2004.04.001 [DOI] [PubMed] [Google Scholar]
- Sugimoto H., Kusumi K., Noguchi K., Yano M., Yoshimura A., Iba K. (2007). The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J. 52 512–527. 10.1111/j.1365-313X.2007.03251.x [DOI] [PubMed] [Google Scholar]
- Tanz S. K., Kilian J., Johnsson C., Apel K., Small I., Harter K., et al. (2012). The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LCHB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings. Plant J. 69 743–754. 10.1111/j.1365-313X.2011.04833.x [DOI] [PubMed] [Google Scholar]
- Terada K., Kanazawa M., Bukau B., Mori M. (1997). The human DnaJ homologue dj2 facilitates mitochondrial protein import and luciferase refolding. J. Cell Biol. 139 1089–1095. 10.1083/jcb.139.5.1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., Froehlich J. E., Koksharova O., Pyke K. A., van Erp H., Osteryoung K. W. (2003). ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15 1918–1933. 10.1105/tpc.013292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P., Bursac D., Law Y. C., Cyr D., Lithgow T. (2004). The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5 567–571. 10.1038/sj.embor.7400172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing A., Williams M. E., Bourett T. M., Hu W., Hou Z., Meeley R. B., et al. (2014). A pair of homoeolog ClpP5 genes underlies a virescent yellow-like mutant and its modifier in maize. Plant J. 79 192–205. 10.1111/tpj.12568 [DOI] [PubMed] [Google Scholar]
- Yang X. Q., Zhang W. W., Li Y., He H. L., Bie B. B., Ren G. L., et al. (2014). High-resolution mapping of the dull fruit skin gene D in cucumber (Cucumis sativus L.). Mol. Breed. 33 15–22. 10.1007/s11032-013-9927-8 [DOI] [Google Scholar]
- Yoo S. C., Cho S. H., Sugimoto H., Li J., Kusumi K., Koh H. J., et al. (2009). Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol. 150 388–401. 10.1104/pp.109.136648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Luo X., Hu B., Wan Y., Xie J. (2013). YGL138(t), encoding a putative signal recognition particle 54 kDa protein, is involved in chloroplast development of rice. Rice 6:7. 10.1186/1939-8433-6-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Li J., Yoo J. H., Yoo S. C., Cho S. H., Koh H. J., et al. (2006). Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol. Biol. 62 325–337. 10.1007/s11103-006-9024-z [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhang D., Han S., Zhang X., Yu D. (2011). Identification and gene mapping of a soybean chlorophyll-deficient mutant. Plant Breed. 130 133–138. 10.1111/j.1439-0523.2010.01844.x [DOI] [Google Scholar]
- Zhou Q., Wang S. H., Hu B. W., Chen H. M., Zhang Z. H., Huang S. W. (2015). An accumulation and replication of chloroplasts 5 gene mutation confers light green peel in cucumber. J. Integr. Plant Biol. 57 936–942. 10.1111/jipb.12355 [DOI] [PubMed] [Google Scholar]
- Zhu J. K., Shi J., Bressan R. A., Hasegawa P. M. (1993). Expression of an Atriplex nummularia gene encoding a protein homologous to the bacterial molecular chaperone DnaJ. Plant Cell 5 341–349. 10.1105/tpc.5.3.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


