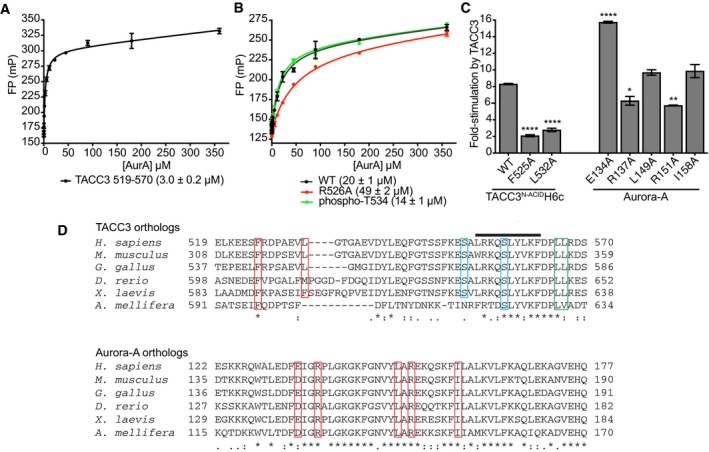
FP assay between FAM‐TACC3 519–570 and Aurora‐A. Reactions were performed in triplicate. Error bars represent standard deviation. The binding affinity between WT Aurora‐A and FAM‐TACC3 519–570 was 3.0 ± 0.2 μM.
FP assays between wild‐type FAM‐TACC3 site 1 peptide (residues 522–536) and mutants, and Aurora‐A. Reactions were performed in triplicate. Error bars represent standard deviation. The binding affinity between WT Aurora‐A and FAM‐TACC3 522–536 was 20 ± 1 μM. In contrast, the K
d for the Aurora‐A interaction with a FAM‐TACC3 522–536 peptide incorporating the R526A mutation was 49 ± 2 μM, a modest increase consistent with the minor contribution of the residue to the interface with Aurora‐A. One further peptide, which incorporated phosphorylation of T534, was determined to have a K
d of 14 ± 1 μM. This peptide was based on the location of a sulphate ion, originating from the crystallization solution, at the interface of the Aurora‐A/TACC3 interface in the crystal structure, close to TACC3 T534. The modest increase in affinity suggests that this post‐translational modification is not significant in regulation of the interaction.
In vitro radioactive kinase assay to determine the effect of TACC3 and phosphorylated Aurora‐A mutants on Aurora‐A kinase activity. Myelin basic protein (MBP) was used as a generic kinase substrate. Incorporation of radioisotope was measured by scintillation counting. Reactions were performed in duplicate. Error bars represent standard error. Assays were compared to the WT TACC3N‐ACIDH6c reaction by one‐way ANOVA with Dunnett's post hoc test. *P < 0.1, **P < 0.01 and ****P < 0.0001.
Multiple sequence alignment of TACC3 (above) and Aurora‐A (below) orthologs spanning the interaction binding interface. Key binding residues are boxed in red. The TACC3 phosphorylation sites, S552 and S558, are boxed in blue. The TACC3 LL motif required for binding to CHC is boxed in green. The TACC3 α‐helix produced on binding to CHC is identified by a black line.