Figure 1. Model for the function of nuclear-encoded PPR and other RNA-binding proteins in protecting plastid RNAs from ribonucleolytic attack.
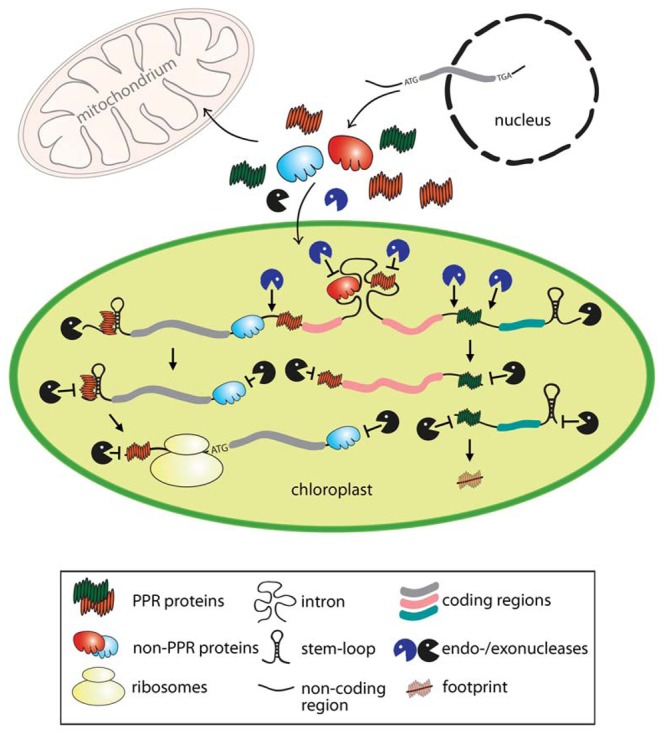
The mode of action shown is representative also for mitochondria. The two colors of protectors (PPR and non-PPR proteins) point to the specificity for particular targets. The scheme shows the endo- and exonucleolytic processing of a representative tricistronic precursor transcript resulting in three stable and translational competent monocistronic products. In chloroplasts, unprotected sites of precursor mRNAs are cleaved by site-specific endonucleases (blue) often in intergenic regions, giving rise to new 5′ and 3′ transcript ends. These extremities are protected against attacks of exonucleases (black) either by the specific binding of RNA protectors and/or by stabilizing secondary RNA stem–loop structures at both ends. For example, PPR10, HCF152, and CRP1 delineated in green show that binding to intergenic regions and subsequent endonucleolytic processing of both adjacent regions results in RNAs with overlapping 5′- and 3′-UTRs, which are protected by the same PPR protein. Binding of protectors to endonuclease-sensitive sites (e.g. intron regions) prevents the cleavage and subsequent unspecific exonucleolytic degradation of the processed RNAs. Furthermore, binding of these factors can stimulate the restructuring of the 5′ region that in turn promotes translation initiation. Short noncoding RNA fragments resulting from the protective role of tightly bound PPR proteins are shown as footprints.
