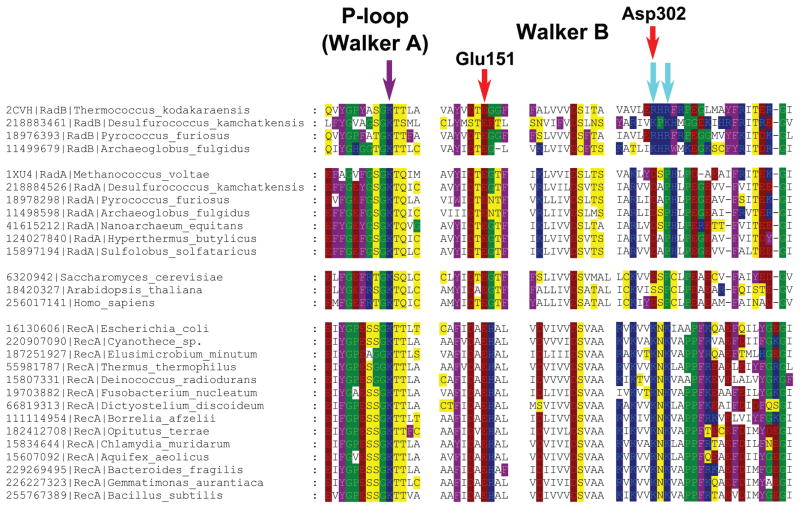
Fig. 4.
Multiple alignment of RecA/RadA/RadB and their Rad51-like eukaryotic homologs. Amino acid residues are colored as in Fig. 1. The violet arrow indicates the P-loop lysine residue, and the two cyan arrows indicate two lysine residues in RecA sequences, which are inserted into the ATP-binding site of the adjacent monomer. Red arrows mark two acidic residues involved in potassium binding in M. voltae (the numbers correspond to the M. voltae sequence). The multiple alignment was constructed using Muscle [147] and visualized with GeneDoc.