Clonal mutations of the TP53, NOTCH1, SF3B1 or BIRC3 genes have allowed to refine the conventional fluorescence in situ hybridization (FISH)-based prognostic stratification of chronic lymphocytic leukemia (CLL) by reclassifying ∼ 20% of good-risk patients (del13q, +12, normal FISH) into higher-risk subgroups.1 Consequently, two novel CLL good-risk subgroups, both devoid of the four gene clonal mutations, have been defined: low risk—harboring +12 or a normal FISH (10-year overall survival (OS): 57%)—and very-low risk— carrying del13q only, whose 10-year OS (69.3%) does not differ from that of the healthy age-matched general population.1
Cytogenetics using novel mitogens allows to recognize chromosomal aberrations in regions uncovered by standard FISH and to identify novel genetic subgroups with prognostic relevance, especially within CLL carrying del17p.2–5 Less is known about the integration of karyotype (KT)-based subgroups into the good-risk CLL categories.
This study was designed to further refine, by KT analysis, the prognostic stratification of CLL with del13q or normal FISH, with the aim of identifying at diagnosis patients who may never require treatment and with a possible normal life expectancy.
Good-risk FISH CLL patients were selected from a series of 248 consecutive untreated patients characterized by FISH and cytogenetics with novel mitogens at diagnosis or within 1 year from diagnosis at two institutions (Rome and Ferrara), as described.2 Dohner’s hierarchic categories were as follows: high risk (del17p/del11q) in 46 cases (18.6%), intermediate risk (+12) in 34 cases (13.7%) and good-risk FISH (del13q only/normal) in 168 cases (67.7%).
Among the 168 good-risk FISH subjects, Sanger sequencing of TP53, NOTCH1, SF3B1 and BIRC3 genes was retrospectively performed in 123 cases with available material. Clonal mutations were identified in 17 patients (13.8%): TP53 in 1 (0.8%), SF3B1 in 8 (6.5%), NOTCH1 in 7 (5.7%) and BIRC3 in 1 (0.8%). KT with novel mitogens was evaluable in 120/123 cases (97.6%): a normal KT or del13q only was present in 83 cases (69.2%), 1–2 lesions (other than del13q) were found in 31 (25.8%) and a complex KT (CKT) in 6 (5%) (Tables 1a and 1b, and Supplementary Figure 1).
Table 1a.
Details of the 30 very-low-risk CLL with 1–2 lesions (other than del13q) or CKT (defined as the detection of three or more chromosomal abnormalities in the same clone)
| KT | UPN | FISH | IGHV | Binet stage | |
|---|---|---|---|---|---|
| 1–2 lesions (27) | 194 | 46,XY,del(6)(q21)[6]/46,XY[14] | Norm | M | C |
| 207 | 46,XY,t(1;13)(p35;q13)[16]/46,XY[1] | 13q- | UM | A | |
| 219 | 47,XXX,−6,+8[3]/46,XX[16] | 13q- | UM | A | |
| 231 | 46,XY,del(14)(q32)[2]/46,XY[18] | Norm | UM | A | |
| 234 | 46,XX,t(1;13)(p31;q21)[14]/46,XX[12] | 13q- | Na | A | |
| 256 | 47,X,add(X)(p11),+del(3)(p11p14)[4]/46,XX[16] | Norm | M | B | |
| 259 | 46,XY,t(13;22)(q14;q12)[6]/46,XY[14] | 13q- | M | A | |
| 274 | 46,XY,del(13)(q14q22),del(14)(q31)[3]/46,XY[17] | 13q- | M | A | |
| 276 | 46,XX,del(14)(q31)[17]/46,XX[17] | 13q- | M | A | |
| 284 | 46,XY,del(13)(q14q31)[10]/46,idem,add(17)(p12)[5]/46,XY[5] | 13q- | M | C | |
| 288 | 46,XY,add(2)(q36)[8]/46,idem,del(14)(q24)[5]/46,XY[7] | Norm | UM | B | |
| 294 | 47,XY,+18[12]/46,XY[8] | Norm | M | A | |
| 299 | 46,XY,del(13)(q14q22)[3]/46,idem,del(14)(q31q32)[4]/46,XY[16] | 13q- | M | A | |
| 303 | 46,XY,del(14)(q32)[2]/46,XY[18] | 13q- | UM | A | |
| 305 | 46,XY,del(7)(q32)[4]/46,XY[21] | Norm | UM | A | |
| 307 | 46,XY,t(4;11)(q21;q24),add(22)(p13)[8]/46,XY,dic(15;15)(p11;p11)[3]/46,XY[9] | Norm | M | B | |
| 315 | 46,XY,t(13;16)(?q12;p13)[8]/46,XY[12] | Norm | M | A | |
| 335 | 46,XY,der(16)t(14;16)(q32;q23)[11]/46,XY[9] | Norm | UM | C | |
| 357 | 46,XY,t(16;18)(q24;q21)[19]/46,XY[1] | 13q- | UM | A | |
| 358 | 46,XY,del(14)(q32)[3]/46,XY[17] | 13q- | M | B | |
| 375 | 46,XY,del(13)(q14)[2]/47,idem,+mar[2]/46,XY[16] | 13q- | M | A | |
| 380 | 46,XY,del(6)(q13)[7]/46,idem,del(2)(q33q37)[4]/46,idem,del(13)(q12q14)[4]/46,XY[6] | 13q- | UM | A | |
| 384 | 46,XY,del(14)(q22q31),i(18)(q10)[16]/46,XY[4] | Norm | M | C | |
| 385 | 46,XX,del(13)(q14q22),del(14)(q32)[10]/46,XY[10] | 13q- | M | A | |
| 393 | 46,XY,t(12;13)(q21;q24)[17]/46,XY[3] | 13q- | M | A | |
| 401 | 46,XX,add(2)(p25)[17]/46,XX[3] | Norm | M | A | |
| 413 | 46,XX,ins(3;2)(p21;p14p16)[8]/46,XX,del(9)(p23)[6]/46,XX[11] | Norm | M | B | |
| ⩾ 3 lesions (3) | 266 | 46,XY,t(14;18)(q32;q21)[5]/46,XY,del(1)(q32),add(9)(q34),−14,+mar[4]/46,XY[12] | Norm | M | A |
| 287 | 46,XY,del(13)(q14q22)[2]/46,idem,−20,+22[13]/46,XY[5] | 13q- | M | A | |
| 309 | 46,XX,del(1)(q23),add(3)(p24),del(13)(q14q22)[10]/46,XX[21] | Norm | M | A |
Table 1b.
Details of the 17 low-risk FISH CLL with gene mutations
| KT | UPN | FISH | IGHV | Mutated gene | Binet stage | |
|---|---|---|---|---|---|---|
| Normal/13q- (10) | 196 | 46,XY[20] | Norm | UM | SF3B1 | A |
| 214 | 46,XX[20] | Norm | UM | SF3B1 | B | |
| 226 | 46,XY[20] | Norm | UM | NOTCH1 | A | |
| 261 | 46,XX,del(13)(q14q22)[4]/46,XX[16] | 13q- | M | SF3B1 | A | |
| 262 | 46,XY[20] | Norm | M | BIRC3 | A | |
| 285 | 46,XX[20] | Norm | UM | SF3B1 | B | |
| 296 | 46,XY,del(13)(q14q22)[5]/46,XY[15] | 13q- | UM | SF3B1 | A | |
| 322 | 46,XY[26] | Norm | M | SF3B1 | B | |
| 336 | 46,XX[20] | Norm | UM | NOTCH1 | A | |
| 396 | 46,XY,del(13)(q14q22)[9]/46,XY[11] | 13q- | UM | SF3B1 | A | |
| 1–2 lesions (4) | 248 | 46,XX,del(7)(q32)[12]/46,XX[10] | Norm | M | SF3B1 | C |
| 338 | 47,XY,+ 18[12]/46,XY[8] | 13q- | UM | NOTCH1 | C | |
| 381 | 46,XY,del(4)(?p12)[2]/46,XY[22] | 13q- | UM | NOTCH1 | B | |
| 390 | 46,XY,t(13;16)(q14;q24)[20] | 13q- | UM | NOTCH1 | B | |
| ⩾ 3 lesions (3) | 191 | 40–47,XY,del(1)(p11),+del(1)(q11),add(2)(p25),+4,−6,+3mar1[cp15]/46,XY[5] | Norm | M | TP53 | C |
| 379 | 46,XY,t(1;14)(p35;q32)[6]/46,idem,der(?)t(?;8)(?;?),−20[7]/46,idem,t(10;13)(?q24;q14)[2]/46,XY[4] | 13q- | UM | NOTCH1 | A | |
| 387 | 43,XY,add(2)(p25),−8,add(11)(p15),−13,der(13)t(13;15)(p13;q15),der(14)t(13;14)(q14;p13),−15,der (16)t(8;16)(q12;p13)[cp19]/46,XY[1] | 13q- | UM | NOTCH1 | A |
Additional chromosomal lesions/CKT (n = 37) were significantly more common, but not exclusive, in patients with more advanced clinical stages—23/98 (23.5%) stage A and 14/22 (63.6%) stage B/C (P = 0.0006)—and significantly associated with the expression of CD38 >30% (P = 0.0038), reinforcing the notion that, in CLL, the genomic complexity arises predominantly in the CD38+ cell fraction.6 Contrariwise, they were not associated to gender or age and equally represented in CLL with del13q only by FISH (21/70, 30%) and normal FISH (16/50, 32%) (P = 0.8). They were more frequent in unmutated (14/36, 38.9%) than in mutated IGHV cases (22/78, 28.2%), though not significantly (P = 0.28).
Overall, a KT with additional chromosomal lesions/CKT was a significant predictor of OS (P = 0.0038—HR = 6.012; 95% CI = 1.78–20.28) and time-to-first treatment (TFT) (P < 0.0001—HR = 4.82; 95% CI = 2.27–10.24) in a univariate analysis including age, gender, CD38, ZAP70, IGHV status, FISH, KT and the four gene mutations, and was an independent predictor of short TFT (P = 0.0004—HR = 5.245; 95% CI = 2.08–13.21)—together with CD38+, unmutated IGHV and normal FISH (Supplementary Table 1 and Supplementary Figure 2).
Comparing the 17 CLL mutated in 1 of the 4 genes with the 103 very-low-risk CLL wild type (WT) for all genes, a normal KT/del13q only was present in 10 (58.8) vs 73 (70.9%) cases, 1–2 lesions (other than del13q) in 4 (23.5) vs 27 (26.2%), CKT in 3 (17.7) vs 3 (2.9%) cases, respectively. CKT was significantly more frequent in the former group (P = 0.036). As expected, the very-low-risk CLL WT showed less-advanced clinical stages (Binet stage A/B/C in 92/9/5 vs 9/5/3 cases, P = 0.0026) and a significantly more benign biologic profile (CD38+: 15.2 vs 64.7%, P = 0.0001; unmutated IGHV genes: 24 vs 70.6%, P = 0.0003) than the CLL with mutations, with no difference in gender or age distribution.
Focusing on the 103 CLL with favorable FISH and WT for clonal mutations, 30 (29.1%) shifted to an intermediate–high-risk category identified by refined KT analysis (Table 1a), regardless of the IGHV gene status (P = 0.44). CLL with del13q by FISH/WT (n = 62) showed additional KT lesions in 25.8% of cases (1–2 lesions in 15, CKT in 1); CLL with normal FISH/WT (n = 44) showed KT lesions in 34.1% of 41 evaluable cases (1–2 lesions in 12, CKT in 2) (P = 0.38). Among them, after a median follow-up of 42.5 months (range, 2.0–102.0), those with normal KT/del13q only showed a significantly longer OS than cases with additional lesions/CKT (P = 0.001), and this was superimposable to the OS of the general age-matched Italian population (Figures 1a and b). Twenty patients (19.4%) underwent treatment; TFT was significantly longer for those with normal KT/del13q than for cases with additional lesions/CKT (P < 0.0001) (Figure 1c).
Figure 1.
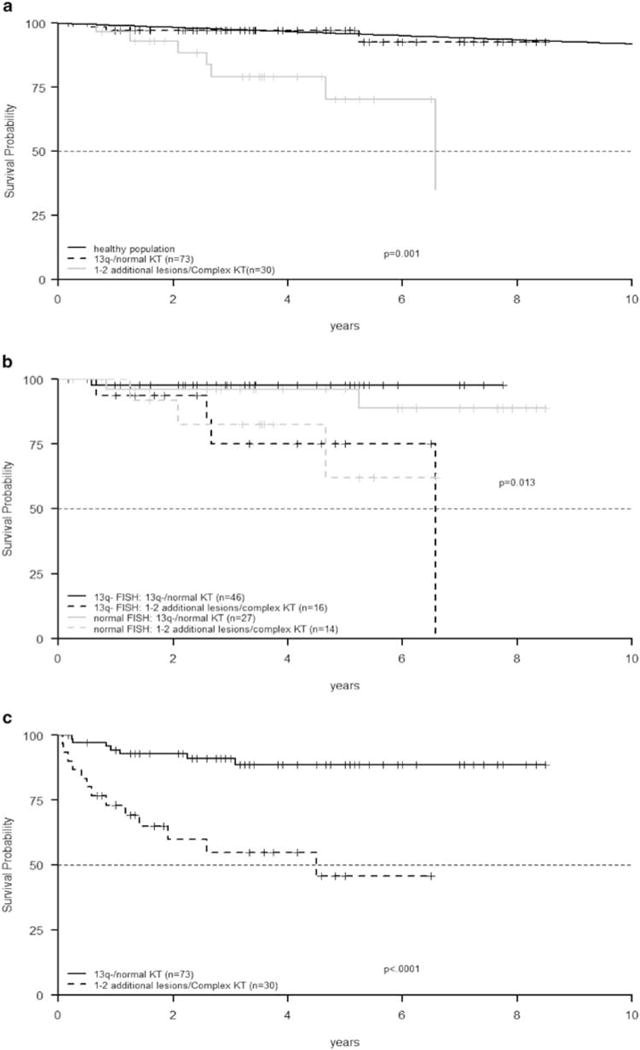
Among good-risk FISH CLL patients devoid of gene mutations, those with normal KT/del13q only were associated with a significantly longer OS than WT cases with additional lesions/CKT (P = 0.001). OS of the general age-matched Italian population was extrapolated by the data on http://www.mortality.org/(a); good-risk FISH CLL patients devoid of gene mutations with normal KT/del13q only were associated with a significantly longer OS than WT cases with additional lesions/CKT even among del13q (P = 0.005) and normal FISH CLL (P = 0.03) subgroups (b); TFT was significantly longer for good-risk FISH CLL devoid of gene mutations with normal KT/del13q than for WT cases with additional lesions/CKT (P < 0.0001) (c).
Subclonal TP53 mutations, evaluated by ultra-deep sequencing on the Genome Sequencer Junior (Roche-454 Life Sciences) (Roche, Mannheim, Germany) and a dedicated bio-informatic analysis,7 were identified in 5/103 very-low-risk CLL (4.8%), with a minor allele frequency from 0.007 to 0.03. Four cases showed a normal KT/del13q and one carried an additional lesion. All are alive and three are still untreated at the last follow-up.
Finally, the six cases with a CKT would have been missed by the analysis of conventional clinico-biologic parameters: five were in stage A, three showed a normal FISH, three a del13q, four had mutated IGHV, two a negative CD38, three were WT for the four genes, two showed NOTCH1 and one had a TP53 mutation. This reinforces the notion that CLL cases with a CKT are not necessarily associated with poor conventional prognostic markers.
Although the first evidence that the number of chromosomal abnormalities detected by classic chromosome banding analysis had prognostic relevance in CLL goes back to 1990, the low proliferative activity of CLL cells in vitro has prevented an extensive application of KT analysis, leading to a four-probe FISH panel to become the cornerstone of CLL prognostic assessment. The subsequent introduction of immunostimulatory CpG-oligonucleotides and IL-28 has enabled to overcome this technical limitation, allowing to obtain an informative KT virtually in all CLL cases and revealing a much more heterogeneous pattern of abnormalities compared with the FISH-based data. A CKT has been significantly associated to CD38 expression and an unmutated IGHV status, with del17p in 28% of cases;8 its frequency increased according to the disease phase, being present in 18.7% of CLL patients at diagnosis and in 30.1% of previously treated patients.9 In line with our observations, CLL with isolated del13q by FISH showed additional lesions/CKT in 34% of cases.8 Among the untreated cases, the number of chromosome aberrations improved the prognostic stratification for OS and TFT based on conventional clinico-biologic features.9
These observations have been more recently confirmed on 1001 CLL cases evaluated within 6 months from diagnosis.10 A CKT (15.7% of cases), significantly associated with unmutated IGHV and aberrations of chromosome 17p, resulted in an independent prognostic factor for shorter TFT together with the unmutated IGHV status and advanced clinical stages. Of the 249 del13q CLL, 19 (7.6%), mostly IGHV mutated, showed a CKT that was associated with a significantly shorter OS compared with similar FISH cases without CKT, supporting our observations.
Only one study has integrated the prognostic impact of the 20 most frequent gene mutations, KT and clinico-biologic parameters in 200 consecutive CLL patients at diagnosis.11 Gene mutations were significantly associated to intermediate–high-risk FISH and CKT (15.4%, 28/195). The CKT, associated to the presence of TP53, ATM and MYD88 mutations, was a significant predictor of both TFT and OS, and resulted independent in a multivariate analysis for OS, along with advanced stage, TP53 mutation/deletion and unmutated IGHV.
Thus, KT complexity aggravates the outcome of CLL across all FISH categories, being present from one-third to more than half of del17p CLL3–5,8,9 and, more surprisingly, in about 14% of normal FISH2 and in 7.6% of del13q CLL.8–10 Most importantly, in the past few years, CKT has proved to be a predictive marker of poor response and short OS with both chemoimmunotherapy12–14 and novel agents,15 independently of TP53 disruption.
In the present study, CLL patients belonging to a good-risk FISH subgroup (del13q/normal), which represent the majority of CLL at diagnosis, were reclassified as intermediate–high-risk in 13.8% of cases because of the presence of one of the four gene mutations (TP53, NOTCH1, SF3B1 and BIRC3). Among the remaining very-low-risk CLL patients with del13q/normal FISH and devoid of unfavorable gene mutations, while TP53 subclonal mutations were rare and of uncertain significance, we showed the presence of a CKT in 3% of cases. Furthermore, we documented the prognostic relevance even of 1–2 additional chromosomal lesions, which were present in 26% of very-low-risk patients. On the other hand, their absence identifies a subgroup of CLL patients with an excellent long-term prognosis, beyond the IGHV status. Thus, as in other hematologic malignancies, the evaluation of conventional KT is relevant also in CLL: beyond the identification of patients poorly responsive to chemoimmunotherapy and BCR inhibitors, it allows to improve the precision of prognostic stratification and counseling of CLL patients at diagnosis, and to identify those who may never require treatment and with a normal/near normal life expectancy across the subgroups defined by FISH and gene mutations.
Supplementary Material
Acknowledgments
The authors thank Associazione Italiana per la Ricerca sul Cancro (AIRC), Special Program Molecular Clinical Oncology, 5 × 1000, Milan, Italy (MCO-10007) (RF, GG); Ricerca finalizzata RF-2011-02349712 (GMR, AC, GG and RF); Finanziamento Medi Progetti Universitari 2013, Sapienza University, C26A13CCJ4 (IDG); Finanziamento Medi Progetti Universitari 2014, Sapienza University, C26A14WWA9 (IDG); Progetti di Rilevante Interesse Nazionale (PRIN) 2015ZMRFEA (RF, GG and AC); FAR (Fondo di Ateneo per la Ricerca) 2013, 2014, 2016 of the University of Ferrara (GMR, AC); EV and ES are supported by AIL (Associazione Italiana contro le Leucemie Linfomi e Mieloma, Ferrara); NIH funding, U54CA209997 and U54 CA193313 (RR); Grant No. KFS-3746-08-2015, Swiss Cancer League, Bern, Switzerland and Grant No. 320030_169670/1 Swiss National Science Foundation, Berne, Switzerland (DR).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Rossi D, Rasi S, Spina V, Bruscaggin A, Monti S, Ciardullo C, et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood. 2013;121:1403–1412. doi: 10.1182/blood-2012-09-458265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rigolin GM, Cibien F, Martinelli S, Formigaro L, Rizzotto L, Tammiso E, et al. Chromosome aberrations detected by conventional karyotyping using novel mitogens in chronic lymphocytic leukemia with ‘normal’ FISH: correlations with clinicobiologic parameters. Blood. 2012;119:2310–2313. doi: 10.1182/blood-2011-11-395269. [DOI] [PubMed] [Google Scholar]
- 3.Delgado J, Salaverria I, Baumann T, Martínez-Trillos A, Lee E, Jiménez L, et al. Genomic complexity and IGHV mutational status are key predictors of outcome of chronic lymphocytic leukemia patients with TP53 disruption. Haematologica. 2014;99:e231–e234. doi: 10.3324/haematol.2014.108365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco G, Puiggros A, Baliakas P, Athanasiadou A, García-Malo M, Collado R, et al. Karyotypic complexity rather than chromosome 8 abnormalities aggravates the outcome of chronic lymphocytic leukemia patients with TP53 aberrations. Oncotarget. 2016;7:80916–80924. doi: 10.18632/oncotarget.13106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu L, Kim HT, Kasar SN, Benien P, Du W, Hoang K, et al. Survival of Del17p CLL depends on genomic complexity and somatic mutation. Clin Cancer Res. 2017;23:735–745. doi: 10.1158/1078-0432.CCR-16-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rigolin GM, Saccenti E, Rizzotto L, Ferracin M, Martinelli S, Formigaro L, et al. Genetic subclonal complexity and miR125a-5p down-regulation identify a subset of patients with inferior outcome in low-risk CLL patients. Oncotarget. 2014;5:140–149. doi: 10.18632/oncotarget.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossi D, Khiabanian H, Spina V, Ciardullo C, Bruscaggin A, Famà R, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123:2139–2147. doi: 10.1182/blood-2013-11-539726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgV(H) status and immunophenotyping. Leukemia. 2007;21:2442–2451. doi: 10.1038/sj.leu.2404935. [DOI] [PubMed] [Google Scholar]
- 9.Haferlach C, Dicker F, Weiss T, Schnittger S, Beck C, Grote-Metke A, et al. Toward a comprehensive prognostic scoring system in chronic lymphocytic leukemia based on a combination of genetic parameters. Genes Chromosomes Cancer. 2010;49:851–859. doi: 10.1002/gcc.20794. [DOI] [PubMed] [Google Scholar]
- 10.Baliakas P, Iskas M, Gardiner A, Davis Z, Plevova K, Nguyen-Khac F, et al. Chromosomal translocations and karyotype complexity in chronic lymphocytic leukemia: a systematic reappraisal of classic cytogenetic data. Am J Hematol. 2014;89:249–255. doi: 10.1002/ajh.23618. [DOI] [PubMed] [Google Scholar]
- 11.Rigolin GM, Saccenti E, Bassi C, Lupini L, Quaglia FM, Cavallari M, et al. Extensive next-generation sequencing analysis in chronic lymphocytic leukemia at diagnosis: clinical and biological correlations. J Hematol Oncol. 2016;9:88. doi: 10.1186/s13045-016-0320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foà R, Del Giudice I, Cuneo A, Del Poeta G, Ciolli S, Di Raimondo F, et al. Chlorambucil plus rituximab with or without maintenance rituximab as first-line treatment for elderly chronic lymphocytic leukemia patients. Am J Hematol. 2014;89:480–486. doi: 10.1002/ajh.23668. [DOI] [PubMed] [Google Scholar]
- 13.Herling CD, Klaumünzer M, Rocha CK, Altmüller J, Thiele H, Bahlo J, et al. Complex karyotypes and KRAS and POT1 mutations impact outcome in CLL after chlorambucil-based chemotherapy or chemoimmunotherapy. Blood. 2016;128:395–404. doi: 10.1182/blood-2016-01-691550. [DOI] [PubMed] [Google Scholar]
- 14.Badoux XC, Keating MJ, Wang X, O’Brien SM, Ferrajoli A, Faderl S, et al. Fludarabine, cyclophosphamide, and rituximab chemoimmunotherapy is highly effective treatment for relapsed patients with CLL. Blood. 2011;117:3016–3024. doi: 10.1182/blood-2010-08-304683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson PA, O’Brien SM, Wierda WG, Ferrajoli A, Stingo F, Smith SC, et al. Complex karyotype is a stronger predictor than del(17p) for an inferior outcome in relapsed or refractory chronic lymphocytic leukemia patients treated with ibrutinib-based regimens. Cancer. 2015;121:3612–3621. doi: 10.1002/cncr.29566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


