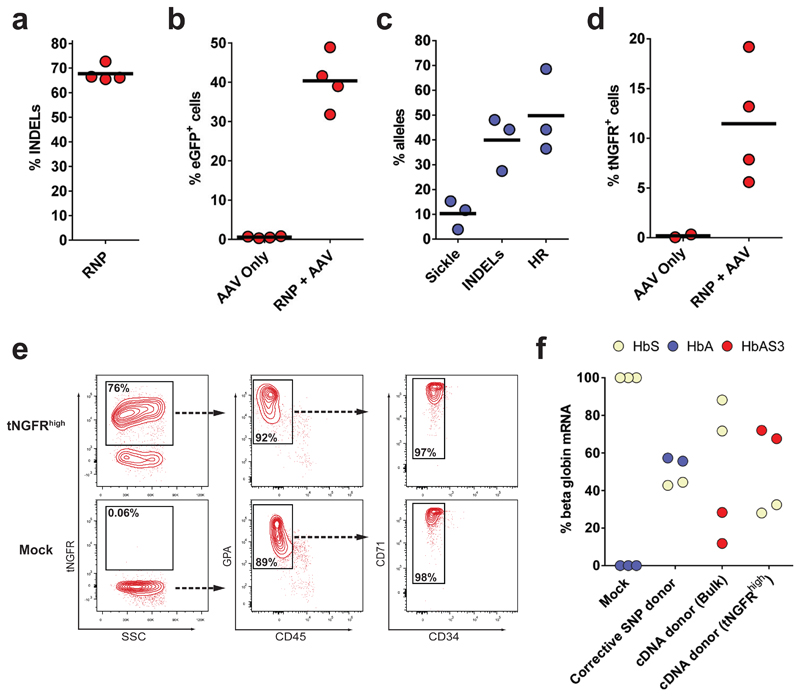
Fig 4. Correction of the E6V mutation in SCD patient-derived HSPCs.
a) gDNA from HBB RNP-treated SCD-HSPCs was harvested and INDELs were analyzed via TIDE software (N=4 different SCD patient donors). b) HBB-targeted SCD-HSPCs were analyzed for GFP expression by flow cytometry (N=4 different SCD patient donors) c) SCD HSPCs were targeted with rAAV6 corrective SNP donor. HBB allele types were analyzed by sequencing of TOPO-cloned PCR fragments derived from In-Out PCR. 50-100 TOPO clones were analyzed from each of three different HSPC donors (N=3 different SCD patient donors). d) SCD HSPCs were targeted with anti-sickling HBB cDNA-EF1-tNGFR correction donor. Frequencies of tNGFR+ cells were analyzed by flow cytometry (N=3 different SCD patient donors). e) SCD Mock HSPCs and sorted SCD tNGFRhigh HSPCs were differentiated into erythrocytes in vitro. Representative FACS plots from Day 21 of differentiation show cell surface markers associated with erythrocytes (GPA+/CD45-/ CD71+/CD34-) f) HbS, HbA, and HbAS3 mRNA expression was quantified by RT-qPCR in erythrocytes differentiated from HBB-edited or Mock SCD HSPCs. All mRNA transcript levels were normalized to the RPLP0 input control (N=2-3 different SCD patient donors).