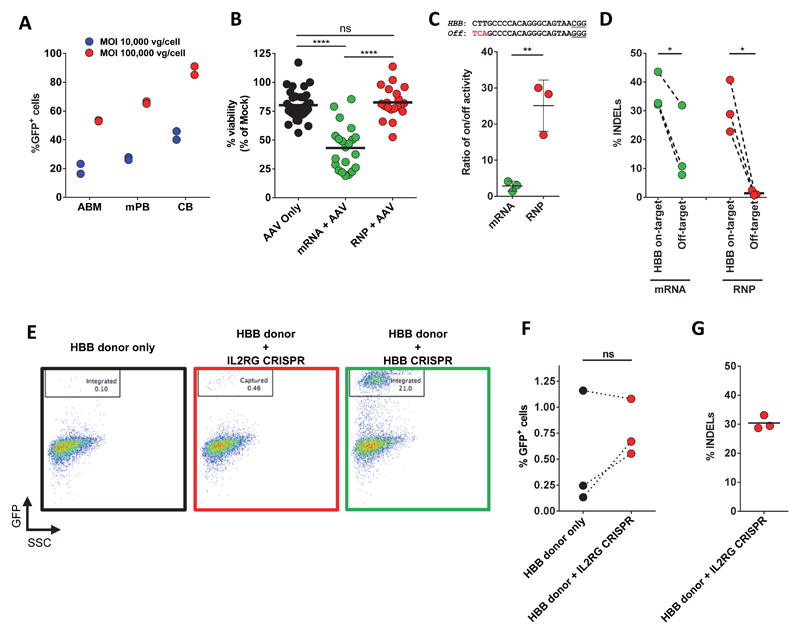
Extended Data Figure 1. High tropism of rAAV6 for CD34+ HSPCs, and viability and specificity assessment of gene editing in CD34+ HSPCs.
(A) CD34+ HSPCs were transduced with a scAAV6 expressing GFP from an SFFV promoter at multiplicities of infections (MOIs) of 10,000 vg/cell or 100,000 vg/cell for 48hrs and then analyzed for percent GFP expression by flow cytometry using a non-transduced sample to set the GFP+ gate at <0.1% GFP+ cells. scAAV was used because it eliminates second strand synthesis as a confounder of actual transduction. Results are from two independent experiments from at least two donors and error bars represent S.D. ABM: Adult Bone Marrow; mPB: Mobilized Peripheral Blood; CB: Cord Blood. (B) CD34+ HSPCs were electroporated with the HBB CRISPR system (mRNA or RNP delivery) or without (AAV only), and then transduced with HBB rAAV6 donor vectors at an MOI of 100,000. Day 4 post electroporation, cells were analyzed by flow cytometry and live cells were gated in high forward scatter (FSC) and low side scatter (SSC). Percent cells in FSC/SSC gate is shown relative to that of Mock-electroporated cells. Each dot represents a unique CD34+ HSPC donor. (C) Upper panel: sgRNA target sequences at the HBB on-target site and a highly complementary off-target site (Chr9:101833584-101833606) are shown. PAM sequences are underlined and red sequence highlights the 3 mismatches of the off-target site. Lower panel: HSPCs were electroporated with either the “All RNA” or RNP-based CRISPR system, and 4 days post electroporation gDNA was extracted and analyzed for INDEL frequencies using TIDE at the on-target HBB and the off-target site. Results are graphed as a ratio of on to off-target activity highlighting the increased specificity of the RNP system. Averages from three different CD34+ HSPC donors are shown and error bars represent S.E.M. ** p < 0.01, **** p < 0.0001, ns = p ≥ 0.05, unpaired Student’s t-test. (D) INDEL frequencies for the data presented in (C) * p < 0.05, paired Student’s t-test. (E) Representative FACS plots showing stable GFP rates at Day 18 post-electroporation in donor-nuclease mismatch experiments. Mismatching nuclease and donor (red box) leads to infrequent end-capture events compared to on-target HR events observed with matched nuclease and homologous rAAV6 donor (green box). HSPCs were electroporated with 15μg Cas9 mRNA and either HBB MS sgRNA or IL2RG MS sgRNA, then transduced with HBB-GFP rAAV6 donor followed by 18 days of culture. (F) End-capture experiments were performed in three replicate experiments each in three unique CD34+ HSPC donors. ns = p ≥ 0.05, paired Student’s t-test. (G) Activity of the IL2RG CRISPR was confirmed by quantification of INDELs at the IL2RG target site using TIDE analysis