Figure 1. Identification of metastatic drivers.
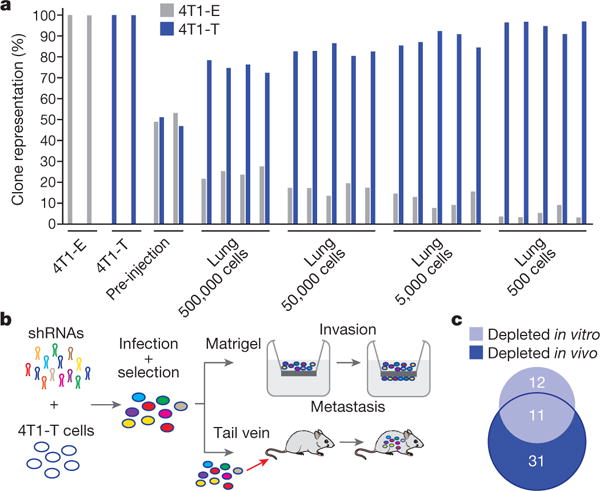
a, Relative proportions of 4T1-E and -T cells extracted from the lungs of NSG mice, into which mixtures of cells were introduced via tail vein at different concentrations. Each bar represents a sample or independent mouse. b, RNAi screening scheme to identify drivers of invasion in vitro and extravasation and colonization in vivo (n = 5 mice or n = 2 Matrigel six-well invasion chambers per approximately 50-construct shRNA pool, gene-level hit calls with empirical Bayes-moderated t-test false discovery rate < 0.05 and 0.1 for in vivo and in vitro screens, respectively). c, Overlap between genes identified in each arm of the RNAi screen depicted in b (hypergeometric test P < 0.01).
