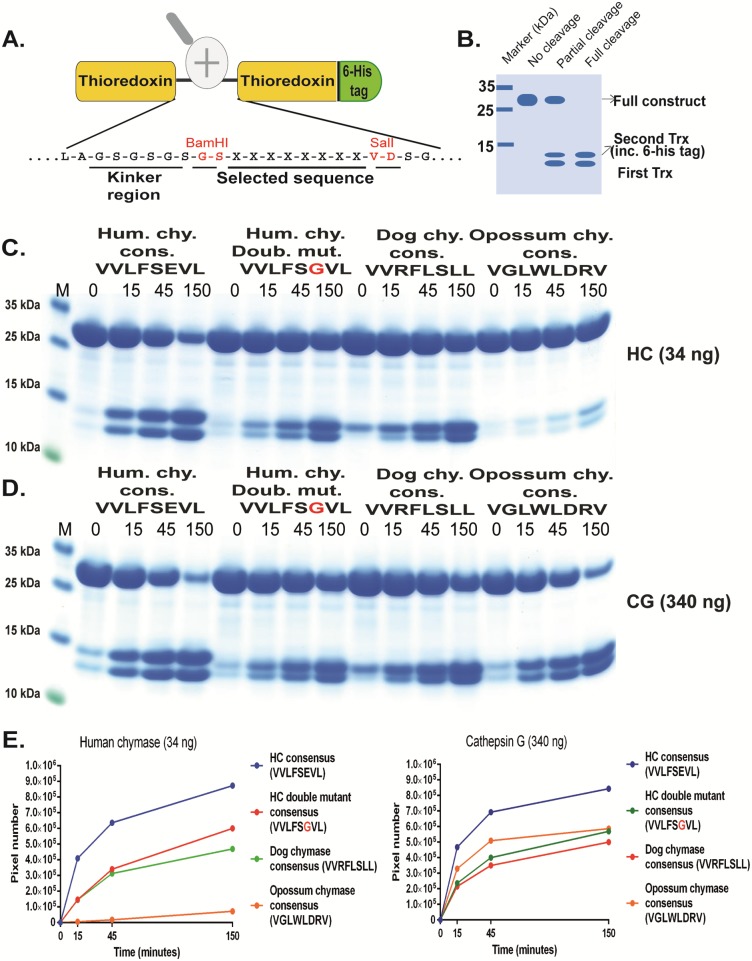
Fig 5. Analysis of the cleavage specificity by the use of recombinant protein substrates.
Panel A shows the overall structure of the recombinant protein substrates used for analysis of the efficiency in cleavage by the hCG and HC. In these substrates two thioredoxin molecules are positioned in tandem and the proteins have a His6-tag positioned in their C termini. The different cleavable sequences are inserted in the linker region between the two thioredoxin molecules by the use of two unique restriction sites, one Bam HI and one SalI site, which are indicated in the bottom of panel A. In panel B a hypothetical cleavage is shown to highlight possible cleavage patterns. Panels C and D shows the cleavage of 4 different substrates by hCG and HC, the HC consensus obtained from phage display, the consensus of a HC double mutant, the dog chymase consensus and the opossum chymase consensus [34, 46, 52]. The name and sequence of the different substrates are indicated above the pictures of the gels. The time of cleavage in min is also indicated above the corresponding lanes of the different gels. The uncleaved substrates have a molecular weight of approximately 25 kDa and the cleaved substrates appear as two closely located bands with a size of 12–13 kDa. In panel E we show the results from scanning of the gels where on e clearly can see the effect of P2´negative charge for both HC and hCG. The gel pictures in panels C and D were densitometrically scanned and the result was presented as two separate diagrams in panel E, one for HC and one for hCG.