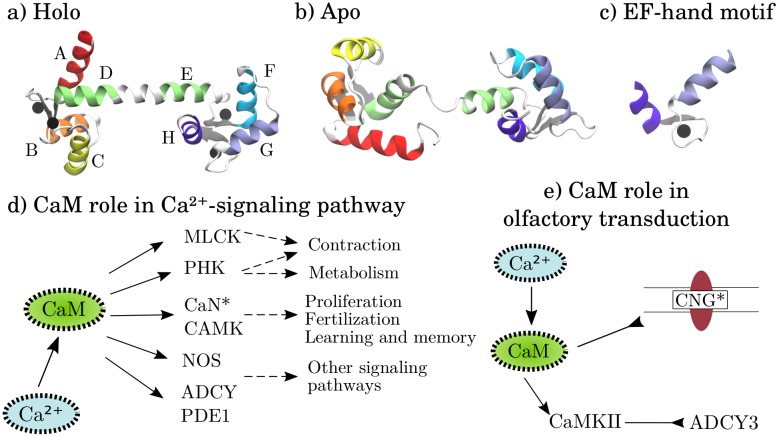
Fig 1. The molecular structure of calmodulin and pathways where calmodulin acts as protein regulator.
Molecular structures of a) holo and b) apo calmodulin. The helices are marked according to their canonical labeling. Ca2+ ions are represented as black spheres and the beta sheets are marked with gray color. c) The EF-hand motif. d) The role of CaM in the Ca2+-signaling pathway. CaM activates the myosin light chain kinase IV (MLCK) and phosphorylase kinase (PHK), calcineurin (CaN), Ca2+/calmodulin-dependent protein kinase (CAMK), nitric oxide synthase 1 (NOS), adenylate cyclase 1 (ADCY) and phosphodiesterase 1A (PDE1). This affects downstream processes such as contraction, metabolism, proliferation, learning etc. e) The role of CaM in olfactory transduction. CaM inhibits the cyclic nucleotide-gated (CNG) channel and activates Ca2+/calmodulin-dependent protein kinase II (CaMKII). CaMKII then inhibits adenylate cyclase 3 (ADCY3). Proteins marked by a star are included in our CaM binding study. The pathways in d) and e) are adapted from KEGG [3].