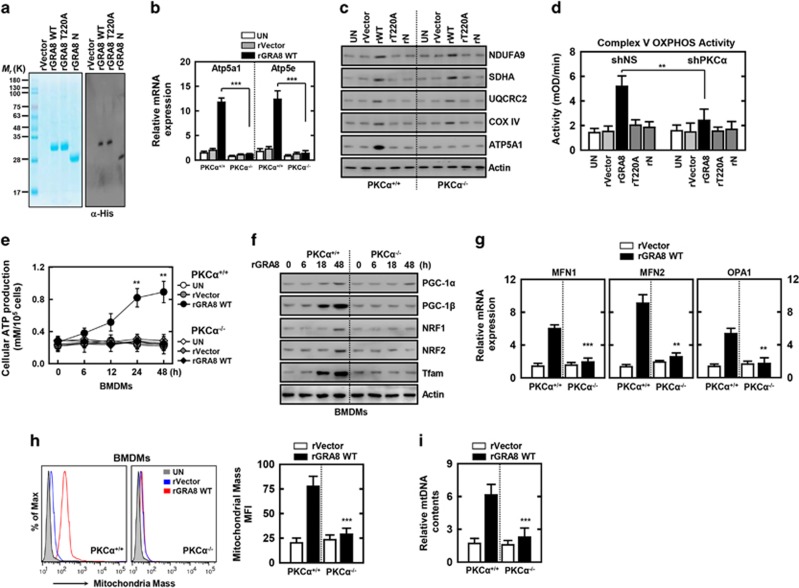
Figure 3.
The rGRA8 treatment increases the induction of mitochondrial activity and biogenesis via protein kinase-Cα (PKCα). (a) Bacterially purified 6xHis-GRA8-WT and its mutants were analyzed by Coomassie blue staining (left) or immunoblotting (IB) with αHis (right). (b–d) Bone marrow-derived macrophages (BMDMs) from PKCα+/+ and PKCα−/− (b, c) or THP-1 cells were transduced with lentivirus-shRNA-NS or lentivirus-shRNA-PKCα (multiplicity of infection (MOI)=100) with polybrene (8 μg ml−1) (right) for 2 days (d), the cells were stimulated with rGRA8 (1 μg ml−1) and its mutants for 6 h (b) and 24 h (c, d) and subjected to quantitative real-time PCR (b), IB (c) or enzymatic activity (d) of oxidative phosphorylation (OXPHOS) genes. (e–g) BMDMs from PKCα+/+ and PKCα−/− were stimulated with rGRA8 for the indicated times and subjected to cellular adenosine triphosphate (ATP) production (e), IB analysis with αPGC-1, αNRF1, αNRF2, αTfam and αActin (f) or quantitative real-time PCR of fusion genes (g). (h) Mitotracker fluorescence signals assessed by a flow cytometric analysis. (Left) Representative histograms from seven independent replicates. (Right) Bar graph indicates the mitochondrial mass mean fluorescence intensities (MFIs). Results are expressed as means±s.d. of seven experiments. (i) Mitochondrial DNA (mtDNA) content in BMDMs measured by quantitative real-time PCR. The mtDNA content was normalized to nuclear DNA. Significant differences (**P<0.01; ***P<0.001) compared with PKCα+/+ or shRNA-NS (Non-specific) (b, d, e and g–i).The data are representative of five independent experiments with similar results (a–g and i).