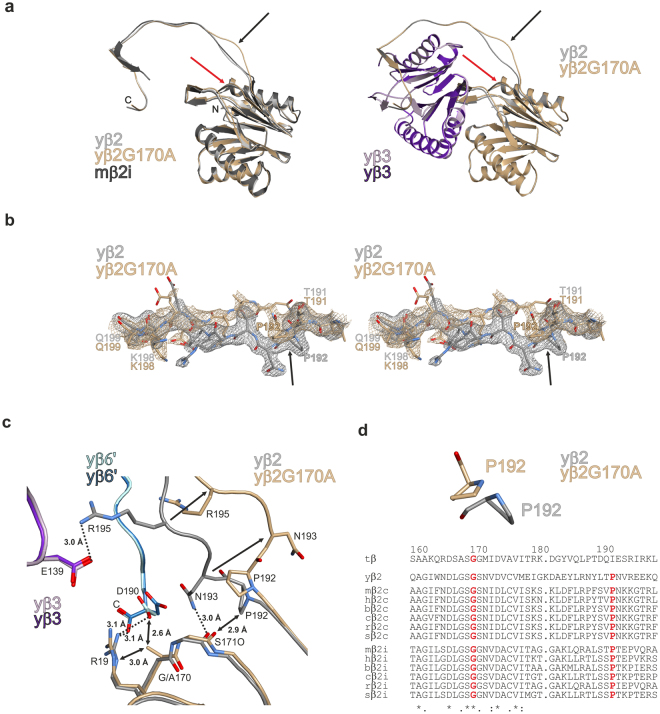
Figure 7.
Crystal structure of the yeast β2-G170A mutant proteasome. (a) Ribbon illustration of the G170A mutant yeast proteasome subunit β2 (brown) superimposed onto the wild type counterpart (grey) and the mouse iCP subunit β2i (black) on the left; association of the subunits β2 and β3 (purple) is shown on the right. The site of mutation and the resulting conformational changes in the β2 C-terminal tail are marked by red and black arrows, respectively. (b) Stereo representation of the 2FO-FC omit electron density maps for the G170A mutant (brownish) and wild type yβ2 subunit (grey) contoured to 1σ. Note the structural changes and the increased flexibility of the loop segment in the mutant structure. The flip of Pro192 is marked by a black arrow. (c) Hydrogen-bonding network around the site of mutation. Exchange of Gly170 in subunit yβ2 results in the reorientation of residues 192–196 (black arrows). Hereby hydrogen bonds (black dotted lines) within yβ2 and with the neighbouring subunits yβ3 and yβ6′ (of the opposite half proteasome) are broken. Steric clashes are marked by black double arrows. (d) The strictly conserved Pro192 is supposed to orient the β2 C-terminus for association with the adjacent β3 subunit. In the mutant crystal structure, flipping of Pro192 changes the conformation of the β2 C-terminus. Species abbreviation in sequence alignment: t: T. acidophilum; y: yeast; m: mouse; h: human; b: Bos taurus; c: Canis familiaris; r: Rattus norvegicus; s: Sus scrofa; amino acid numbers are assigned according to the β-subunit of T. acidophilum.