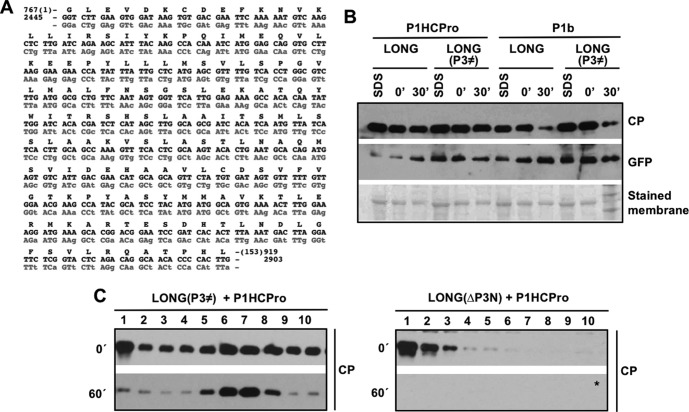
FIG 5.
Amino acid sequence of P3 rather than a particular nucleotide sequence is required for CP stability and proper assembly of virion-like particles. (A) Recoding of the first 153 amino acids of PPV P3 with synonymous mutations. A fragment of the P3 wild-type coding sequence (black) is aligned against the recoded P3 version (gray). The introduced synonymous mutations are indicated in lowercase. Amino acids (black, one-letter code) are indicated at the top of the nucleotide sequence. Nucleotide and amino acid positions using PPV-NK-GFP (EF569215.1) as reference are indicated. (B) Analysis of the stability of the CP produced by agroinfiltration of N. benthamiana leaves with the indicated pLONG mutants along with other constructs. Samples were collected at 5 days postagroinfiltration. Extracts prepared in SDS buffer (SDS) and the time, in minutes, elapsed after extraction with phosphate buffer (0′ and 30′) are indicated. Protein accumulation was assessed by immunoblot analysis with polyclonal serum specific for PPV CP and a monoclonal antibody specific for GFP. The membrane stained with Ponceau red showing the RubisCO large subunit was included as a loading control. (C) Sucrose gradient centrifugation of extracts of N. benthamiana leaves agroinfiltrated with the indicated constructs. Extracts, prepared in phosphate buffer, were loaded on the gradient immediately after being prepared (0′) or after 60 min of incubation at 25°C (60′). Sixteen fractions were collected from the bottom, and the 10 upper ones were subjected to CP-specific immunoblot analysis. The asterisk indicates that the membrane was overexposed.