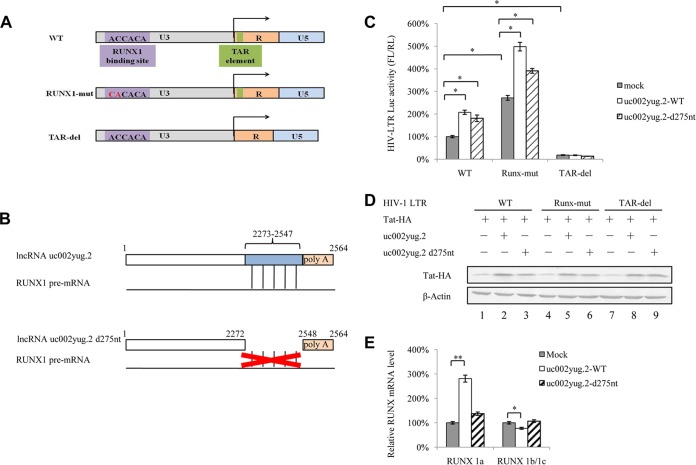
FIG 7.
uc002yug.2 affects HIV-1 transcription through altering RUNX1 and Tat expression. (A) Schematic of HIV-1 LTR mutants. HIV-1 LTR mutants that had previously been shown to lack RUNX binding ability (middle row) or have a deletion in the TAR element (bottom row) were constructed. (B) Schematic of the uc002yug.2 mutant. Compared with the wild-type uc002yug.2 (uc002yug.2-WT), uc002yug.2-d275nt was constructed to delete 275 nt at its 3′ terminus, which is responsible for the alternative splicing of RUNX1 via binding with RUNX1 pre-mRNA (lower diagram). (C) Effect of uc002yug.2 and its mutants on the HIV-1 LTR as well as its mutants. Wild-type or mutant HIV-1 LTR, pRenilla plasmid plus control vector, or uc002yug.2-WT or uc002yug.2-d275nt was cotransfected into HEK293T cells. After 48 h, cells were harvested. The LTR activity was determined by a dual-luciferase reporter assay, and the luciferase activity of HEK293T cells transfected with wild-type HIV-1 LTR, Tat plus control vector, was set as 100%. (D) Tat protein levels in cells were determined by immunoblotting. (E) The mRNA levels of RUNX1a and RUNX1b and -1c in uc002yug.2-WT-, uc002yug.2-d275nt-, and mock-transfected HEK293T cells were detected by qRT-PCR, and the corresponding value of the mock control was set as 100%. Results are representative of those from three independent repeats. Data are presented as means ± SDs.