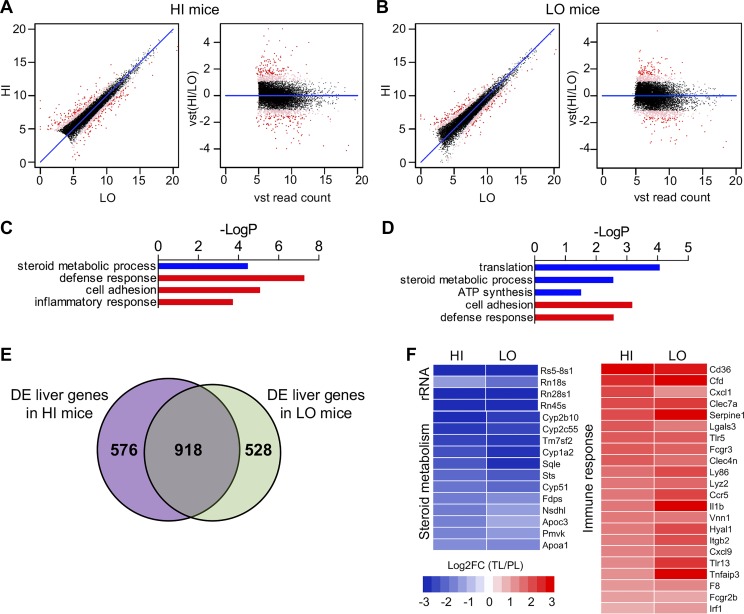
Fig. 7.
Inflammatory genes are induced and ribosomal RNA genes are repressed in liver with the development of diet-induced obesity. XY and VST scatterplots show over 1,400 DE genes (red dots) in a comparison of expression levels between primary (PL) and terminal liver (TL) in the HI group (A) and the LO group (B). C, D: GOrank analysis shows inflammatory or defense response genes were highly upregulated in both HI and LO mice following high-fat feeding. In contrast, the expression of genes related to ribosomal RNA, lipid biosynthesis, and mitochondrial oxidative phosphorylation was reduced in the TL after HFD. E: there was a high degree of similarity between the DE genes of the HI and LO groups with 918 genes shared by both, again predominantly reflecting inflammatory response pathways. F: heat maps show decreased expression of rRNA and steroid metabolism genes and increased expression of immune response genes in liver tissue of both high and low resistance mice following HFD. Please note that relative expression is expressed as the log2-transformed fold change of TL vs. PL mice [or log2FC (TL/PL)].