FIGURE 2.
Organization and molecular structure of the mammalian circadian system
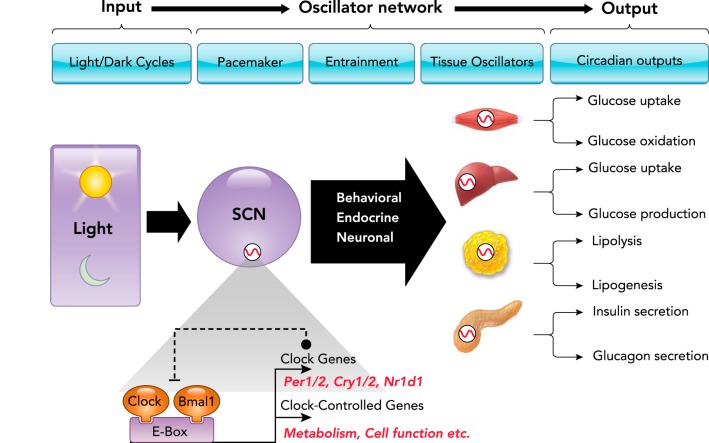
To provide efficient coordination of circadian timing, the mammalian circadian system is organized as a multi-level hierarchical oscillator network. Changes in the LD cycle are perceived by specialized ganglion cells in the retina, synchronizing the central pacemaker of the circadian system in the SCN to the solar day. The main function of the SCN clock appears to coordinate and synchronize cell-autonomous circadian oscillators present in a wide array of peripheral tissues (e.g., skeletal myocytes, hepatocytes, adipocytes, and pancreatic islet cell subtypes) via a combination of neuronal, humoral, and behavioral cues, thus integrating a complex multi-level hierarchical oscillator network. The molecular make up of circadian oscillators consists of the transcriptional activators CLOCK and its heterodimer BMAL1, along with repressor genes that encode period (PER1, 2) and cryptochrome (CRY1, 2) proteins. This regulatory mechanism ensures the generation of 24-h cycles of transcription and translation. Specifically, the CLOCK: BMAL1 heterodimer is essential for the generation of circadian rhythms of transcription through DNA binding to conserved promoter regions (E-boxes) of numerous clock-controlled genes critical for regulation of diverse cellular functions and metabolic pathways.