Fig. 3.
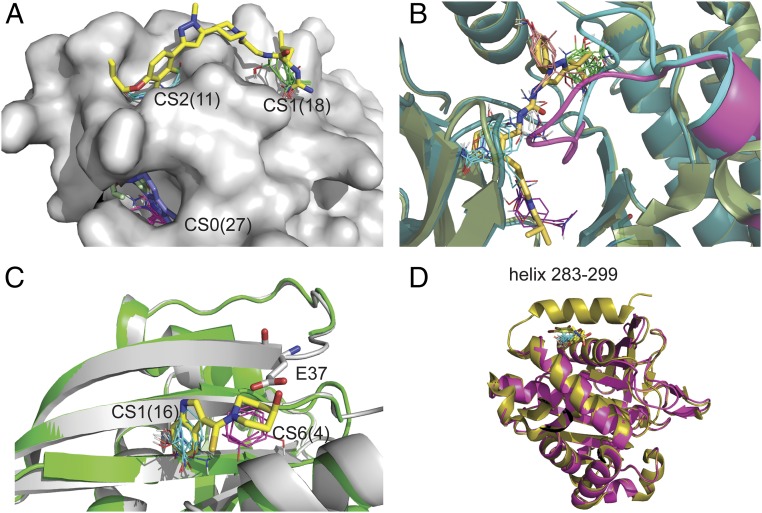
Missing regions in X-ray structures as predictors of cryptic sites. (A) Mapping of the bound IL-2 structure 1PY2 with inhibitor (yellow) shown for reference. The loop of residues 75 to 80 is not visible in the X-ray structure, and the strongest hot spot, CS0 (magenta, 27 probe clusters), is at the gap created by the missing loop, which indicates a cryptic site. The site binds an allosteric ligand (blue) in the structure 1NBP. Other structures also show that the region at this cryptic site is very flexible. For example, in the inhibitor-bound structure 1M48, residues 79 to 82 are missing, but F78 (colored light green) protrudes into the pocket. (B) Substrate binding region of P38 protein kinase. Shown are the unbound structures 3D83 (light green), with the DFG loop shown in magenta, and 2ZB0 (dark blue), with the DFG loop shown in cyan. Both loops are in DFG-in conformation and would clash with the ligand (shown as yellow sticks), superimposed from the bound structure 2YIW, which is in DFG-out conformation. Mapping the ligand-bound DFG-out structures 1KV1, in which residues 171 to183 are missing, yields the hot spots CS0 (green, 23 probe clusters), CS1 (magenta, 12 probe clusters), CS3 (orange, nine probe clusters), and CS4 (white, eight probe clusters). In contrast, mapping the DFG-in structure 2ZB0 yields only the hot spot CS0 (cyan, 16 probe clusters). As shown, mapping both DFG-in and DFG-out conformations, the hot spots map out the entire ligand binding site. (C) Inhibitors of the KRAS/SOS interaction. Shown are chain A (gray) and chain C (green) of the unbound structure 3GFT. Residues 36 and 37 are not visible in chain C. Mapping of chain A identifies the hot spot CS1(cyan, 16 probe clusters) in a hydrophobic pocket that binds the indole group of an inhibitor. The mapping of chain C finds the additional hot spot CS6 (magenta, 4) in a secondary site that enables the binding of the slightly higher affinity inhibitor (yellow sticks) in the structure 4EPW. However, in chain A, the side chain of Glu37 (shown as white sticks) protrudes into this secondary site and would clash with the inhibitor. (D) Protein tyrosine phosphatase 1B (PTP1B). Shown are the unbound structures 1SUG (gold) and 2HNP (magenta), the latter missing the C-terminal helix. 1SUG was mapped after removal of residues 283 to 299, resulting in the hot spot CS1 (cyan, 17 probe clusters), overlapping with the allosteric inhibitor superimposed from the structure 1T48 (yellow sticks).