Significance
Triple-negative breast cancer (TNBC) is responsible for significant mortality among breast cancer subtypes, with its treatment largely unsuccessful due to ineffective targeted therapies. Our bioinformatics analysis demonstrates a unique alternative splicing pattern in TNBC compared with those of other breast cancers. In analyzing the underlying mechanism of the distinct alternative splicing profile, TDP43, a critical gene previously implicated in neurodegenerative disease, is found to promote TNBC progression. Mechanistically, TDP43 regulates extensive alternative splicing events, including downstream gene PAR3, by forming a complex with SRSF3 to regulate alternative splicing events coordinately. Splicing factors TDP43 and SRSF3, which are likely responsible for the unique alternative splicing, could serve as potential targets for TNBC therapy.
Keywords: breast cancer, alternative splicing, TDP43, SRSF3, PAR3
Abstract
Aberrant alternative splicing has been highlighted as a potential hallmark of cancer. Here, we identify TDP43 (TAR DNA-binding protein 43) as an important splicing regulator responsible for the unique splicing profile in triple-negative breast cancer (TNBC). Clinical data demonstrate that TDP43 is highly expressed in TNBC with poor prognosis. Knockdown of TDP43 inhibits tumor progression, including proliferation and metastasis, and overexpression of TDP43 promotes proliferation and malignancy of mammary epithelial cells. Deep sequencing analysis and functional experiments indicate that TDP43 alters most splicing events with splicing factor SRSF3 (serine/arginine-rich splicing factor 3), in the regulation of TNBC progression. The TDP43/SRSF3 complex controls specific splicing events, including downstream genes PAR3 and NUMB. The effect of reduced metastasis and proliferation upon the knockdown of TDP43 or SRSF3 is mediated by the splicing regulation of PAR3 and NUMB exon 12, respectively. The TDP43/SRSF3 complex and downstream PAR3 isoform are potential therapeutic targets for TNBC.
Alternative splicing (AS) is a critical process during posttranscription in which exons from a single gene are assembled in different ways to produce several protein isoforms in eukaryotic organisms (1). Different tissue types exhibit their own splicing pattern characteristics (2). Among breast cancer subtypes, the basal-like subtype, which is mainly parallel with triple-negative breast cancer (TNBC), is characterized by the expression profile of the mammary basal cells (3). Currently, however, there are no effective targeted therapies for such cancers, with chemotherapy being the only established therapeutic option to date (4). Based on expression and mutation profiles, several molecular targets, such as high frequency of TP53 mutation, loss of RB1 and BRCA1, PI3K-pathway activation, and hyperactivated cMYC (5), exhibit promising clinical applications (6). Thus far, however, application of these therapies has been largely unsuccessful due to poor outcomes in clinical trials. Therefore, additional molecular signatures of TNBC need to be identified for improved diagnosis and treatment. As different cell types exert cell-specific splicing patterns, we hypothesized that TNBC may exhibit particular splicing signatures which could bring about new strategies for TNBC treatment.
TDP43 (TAR DNA-binding protein, also named “TARDBP”) is a splicing factor belonging to the hnRNP family, with two RNA recognition motifs (RRMs) and a glycine-rich domain (7). As a RNA-binding protein, TDP43 is involved in the regulation of many biological processes, including transcriptional repression (8), mRNA splicing (9), RNA translocation (10), miRNA processing (11), and mRNA stability (12). Most previous reports on TDP43 have focused on its pathogenesis in amyotrophic lateral sclerosis and frontotemporal lobar degeneration, which are accompanied by abnormally high expression, protein aggregation, phosphorylation, and ubiquitylation (13–15). However, little attention has been devoted to the role of TDP43 in tumor progression.
During the regulation of AS, serine/arginine (SR)-rich proteins are critical components of the machineries of both constitutive and alternative pre-mRNA splicing. Like other members of the SR protein family, SRSF3 contains one N-terminal RNA-binding domain and a downstream SR-rich domain. Previous studies have identified SRSF3 as a proto-oncogene in several types of cancer (16–20) and as a regulator of AS for HER2 splice variants in breast cancer cells (21). The delineation of SRSF3 in breast cancer progression, especially in TNBC, would shed light on the roles of AS in TNBC.
There has been growing interest in the characterization of the roles of splicing factors in the regulation of AS. Recently, various principles of AS have been described (22–24), with the highly context-dependent and position-sensitive regulation of AS being the best-characterized principles (22). Despite these findings, many critical problems remain poorly described. Although hundreds of splicing factors are well known to be involved in specific splicing events (25), how splicing factors, especially for non-small nuclear ribonuclear proteins (snRNPs), regulate AS in coordination remains unclear. Previous reports have investigated the coordinated actions of two splicing factors (21, 26); however, further studies are still necessary to reveal the interaction between two splicing factors in splicing regulation and its role in biological function and disease treatment.
Here, we demonstrate a unique splicing pattern in basal-like breast cancer, which was unlike that of other breast cancer subtypes. As an important member of the splicing factor complex meditating this pattern, TDP43 is highly expressed in TNBC, and loss of its expression suppresses tumor progression both in vitro and in vivo. We found that TDP43 acted in concert with another splicing factor, SRSF3, to regulate a set of AS events in TNBC. Importantly, we identified the downstream target, a PAR3 splicing isoform with a deletion of exon 12, which exerts a role opposite that of its full-length form for migration/invasion of TNBC. Our data identified a splicing factor complex which may be the underlying mechanism for the unique TNBC AS profile and identified TDP43 and PAR3 isoforms with exon 12 deletion as potential therapeutic targets for TNBC diagnosis and drug design.
Results
TDP43 Is a Major Regulator of the Unique AS Profile of TNBC.
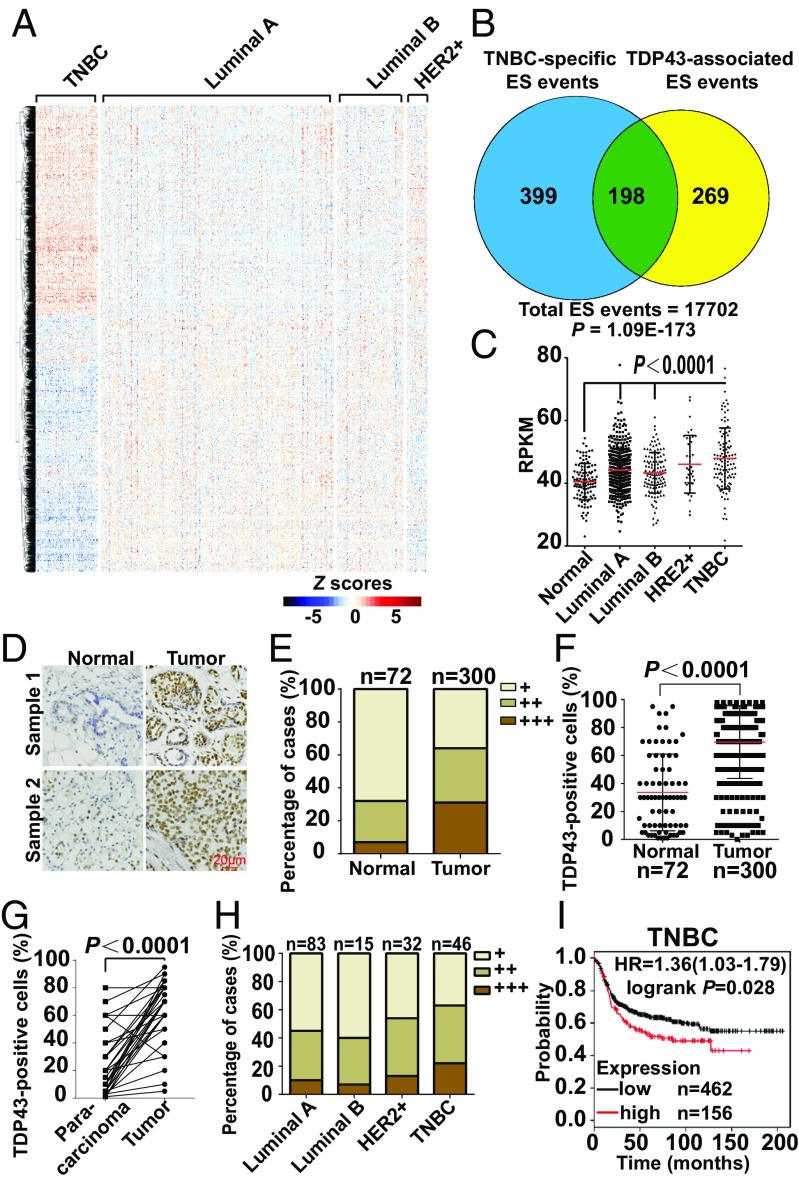
To investigate AS patterns among various breast cancer subtypes, we downloaded percent spliced in (PSI) values for breast cancer splice events from The Cancer Genome Atlas (TCGA) SpliceSeq, a web-based resource (27). A total of 45,421 splice events from 10,480 expressed genes were acquired from 1,094 available samples representing four breast cancer subtypes (luminal A, luminal B, HER2-enriched, and TNBC) (Dataset S1). When the PSI values of splicing events from each breast cancer subtype were compared against a pool of the other three subtypes, TNBC tumors possessed many more significant splicing events (Student’s t test P < 0.001) (Fig. S1A), suggesting a distinctive splicing profile of TNBC compared with the other three subtypes. To show the unique profile of TNBC visually, the 3,049 splicing events with significant differences [|ΔPSI| >0.1, Mann–Whitney test followed by false-discovery rate (FDR) correction, P < 0.05] between any two of the four groups were demonstrated as a heatmap (Fig. 1A) in which TNBC showed a distinct splicing pattern in comparison with the other breast cancer subtypes.
Fig. 1.
Unique alternative splicing profile and high expression of TDP43 with poor prognosis in TNBC. (A) Heatmap of Z scores of PSI values for differential splicing events across breast cancer subtypes. (B) Venn diagram showing the overlap between TNBC-specific ES events and TDP43-associated ES events. The P value (1.09e-173) was calculated by Fisher’s exact test. (C) Transcript expression levels of TDP43 in normal breast tissue and different breast cancer subtypes in the TCGA. RPMK, reads per kilobase of transcript per million reads mapped. (D and E) Immunohistochemical staining (D) and quantification of percentage of cases (E) of TDP43 in tissue chips with normal breast tissue and breast carcinomas. (F) Quantification of the percentage of positively stained TDP43 protein nuclei in grouped samples. (G) Quantification of the percentage of cases of TDP43 in breast cancer and para-carcinoma tissues by immunohistochemical staining of TDP43 in tissue chips. (H) Quantification of the percentage of cases of TDP43 in breast cancer subtypes by immunohistochemical staining of TDP43 in tissue chips. (I) Kaplan–Meier plots showing survival analysis of TNBC subtypes, which were stratified by the expression level of TDP43. HR, hazard ratio.
Splicing factors are critical for the regulation of splicing outcomes. Thus, we determined which splicing factors corresponded to the unique splicing pattern of TNBC. Because splicing factors form complexes in a coordinated manner for AS regulation (22, 28–30), we first identified the coordinately expressed splicing factors using coexpression network analysis. A cluster of 57 coexpressed splicing factors was identified (R = 0.64 ± 0.11; P < 2.55e-12) (Fig. S1B) within 221 known or putative splicing factors (31) using the TCGA breast cancer dataset. Most of the 57 splicing factors exhibited higher expression in TNBC than in non-TNBC subtypes (Fig. S1C), showing significant enrichment in the differentially expressed splicing factors of TNBC within the gene set (43 of 57 vs. 117 of 221, P = 0.002, Fisher’s exact test). Physical interaction network analysis based on protein–protein interaction study (32) confirmed the extensive interactions among the 57 splicing factors (Fig. S1D).
To explore the major splicing factor responsible for the unique profile of TNBC, we evaluated how much of the TNBC-specific splicing profile could be explained by each splicing factor. Among the 57 splicing factors, TDP43-associated events possessed the most significant overlap (P = 1.09e-173, Fisher’s exact test) (Dataset S2), and the overlapped events explained 33% (198 of 597) of TNBC-specific exon-skip (ES) events (Fig. 1B). Subsequent enrichment analysis (Experimental Procedures) also demonstrated that TDP43-associated events had the most significant enrichment of TNBC-specific events (P = 2.20e-87, Fisher’s exact test) (Dataset S2). To further verify the important role of TDP43 in regulating the TNBC splicing pattern, we examined the potential TDP43-binding motif around the cassette exon regions of TNBC-specific ES events. The rMAPS webserver (33) showed that the TDP43-binding motif exhibited significant enrichment downstream of the TNBC-specific cassette exons (P = 3.24e-4, Wilcoxon’s rank sum test) (Fig. S1E).
Taken together, our results suggested that TDP43 played an essential role in the regulation of the unique splicing profile of TNBC.
TDP43 Is Highly Expressed in Breast Tumor with Poor Prognosis.
When analyzing the TCGA database, the TDP43 expression level was found to be significantly higher in breast tumor samples than in normal breast tissue (Fig. 1C). To confirm the expression profile in clinical samples, we immunohistochemically stained tissue chips, which demonstrated markedly higher levels of TDP43 in tumor samples (Fig. 1D). Most tumor samples showed higher enrichment (Fig. 1E), with an average of 70% of TDP43-positive cells in tumor samples compared with 30% in normal samples (Fig. 1F). This result was also confirmed in paired tumor and para-carcinoma tissues (Fig. 1G and Table S2). Interestingly, according to the TCGA database, expression levels were higher in the TNBC subtype than in other subtypes (Fig. 1C). We also evaluated TDP43 expression in tissue chips with subtype classification. Results demonstrated that the expression of TDP43 was higher in TNBC (Fig. 1H and Fig. S1F), which was confirmed in various breast cancer cell lines at both the protein and mRNA levels (Figs. S1G and S3F). Moreover, survival analysis of basal subtypes demonstrated poorer survival probability in patients with higher TDP43 expression (Fig. 1I and Fig. S1H), whereas this was not observed in luminal types. Collectively, these results showed that TDP43 was highly expressed in TNBC with poor prognosis.
Knockdown of TDP43 Inhibits Breast Cancer Progression.
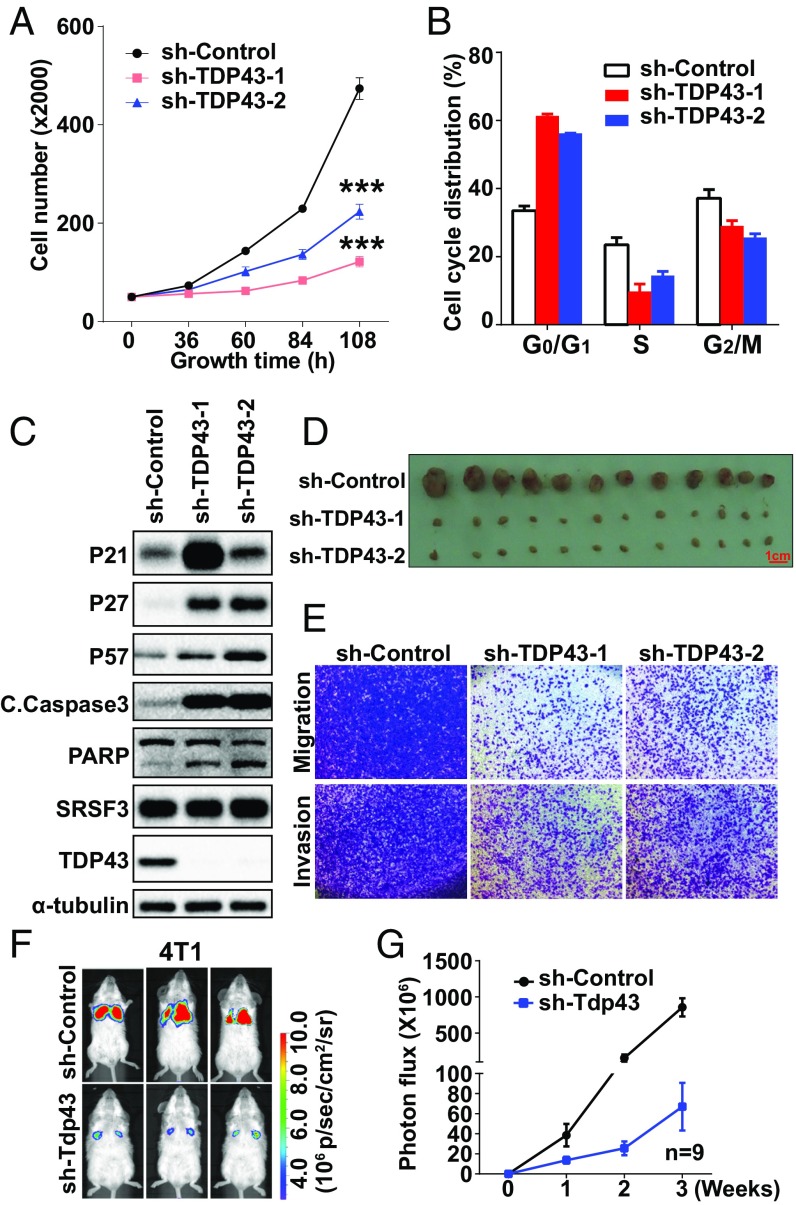
To determine the roles of TDP43 in tumor progression, we investigated cell-proliferation ability after disruption of the TDP43 transcript by shRNA lentivirus in two TNBC cell lines, MDA-MB231 and HCC1806 (knockdown efficiencies are shown in Fig. S2 A and B). In MDA-MB231 cells, knockdown of TDP43 by two shRNAs reduced cell growth in a time-dependent manner, as demonstrated in the cell-growth curve (Fig. 2A). The shRNA with better knockdown efficiency, sh-TDP43-1, showed a greater ability to inhibit cell growth than sh-TDP43-2, the less efficient shRNA. These results were consistent with those obtained from the HCC1806 cell line (Fig. S2C). The BrdU assay revealed that knockdown of TDP43 by both shRNAs decreased the BrdU-positive signals (Fig. S2D). Moreover, loss of TDP43 caused cell-cycle arrest at the G1 phase (Fig. 2B), induced cell apoptosis (Fig. S2 E and F), and promoted the levels of cell-cycle inhibition proteins (P21 and P27) and proapoptotic markers [cleaved caspase-3 and poly (ADP-ribose) polymerase (PARP)] (Fig. 2C). A xenograft assay was performed to examine the effect of sh-TDP43 on tumor growth in vivo. Stable knockdown of TDP43 remarkably reduced tumor volume (Fig. 2D). A Transwell assay demonstrated that MDA-MB231 cell migration and invasion ability decreased upon sh-TDP43 treatment in comparison with the control group (Fig. 2E). To confirm metastasis ability, we injected mouse breast cancer 4T1 cells expressing sh-Tdp43 together with the luciferase gene into the tail vein of mice. Compared with the control group, the mice that received a tail-vein injection of Tdp43-knockdown cells developed fewer lung metastases (Fig. 2 F and G). These data suggested that knockdown of TDP43 inhibited the progression of TNBC.
Fig. 2.
Knockdown of TDP43 inhibits breast cancer progression. (A) Cell-growth inhibition upon TDP43 knockdown by shRNAs. (B) Cell-cycle analysis after knockdown of TDP43. (C) Western blotting of cell-cycle and apoptosis-related proteins. (D) Tumor size after 2 mo of mammary fat pad growth in the xenograft assay. Each experimental group contained six mice. (E) Cell migration and invasion levels by Transwell assays. (F and G) Fluorescence density (F) and statistical analysis (G) of metastasis ability by tail-veil injection using 4T1 cells expressing luciferase (n = 9). Data in A–D are shown as the average ± SEM for three independent measurements; ***P < 0.001. All data are for MDA-MB231 cells, except where labeled.
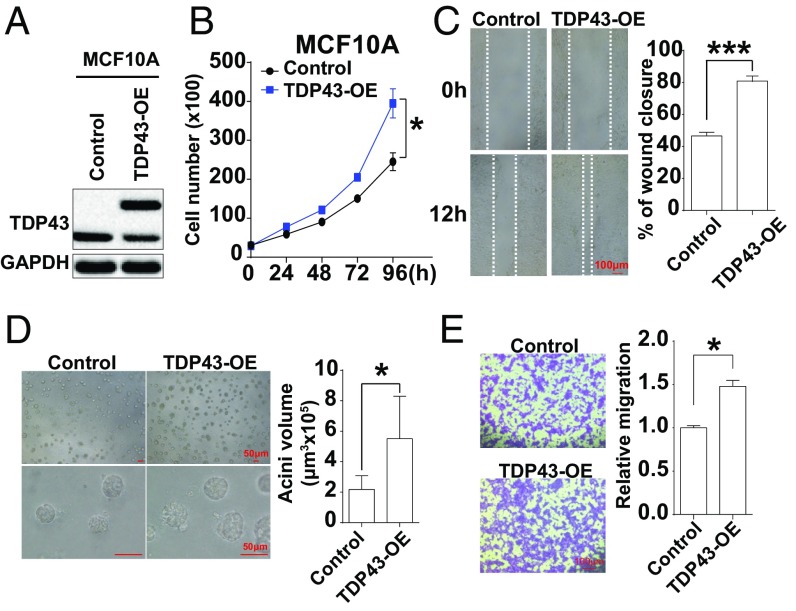
Overexpression of TDP43 Promotes Mammary Epithelial Cell Proliferation and Malignancy.
To further study the role of TDP43 as a pro-oncogene, we stably overexpressed TDP43 in the immortalized MCF10A cell line (Fig. 3A), which is estrogen receptor (ER)-, progesterone receptor (PR)-, and HER2-negative. Ectopic overexpression of TDP43 promoted cell proliferation in a time-dependent manner (Fig. 3B). The 3D cell culture revealed that MCF10A cells overexpressing TDP43 developed larger acini in than the cells in the vehicle group, indicating more potent malignancy with TDP43 overexpression (Fig. 3C). Moreover, the wound-healing assay showed a significant rise in cell motility following the up-regulation of TDP43 expression (Fig. 3D). The Transwell assay also demonstrated that overexpression of TDP43 induced cell migration in comparison with the control group (Fig. 3E). These results indicated that TDP43 promotes breast cancer progression, consistent with the phenotypes observed with the loss of TDP43 in TNBC cells.
Fig. 3.
Overexpression of TDP43 promotes mammary epithelial cell proliferation and malignancy. (A) TDP43 protein levels in MCF10A cells expressing Flag-TDP43 or vehicle control by Western blotting. (B) Cell growth increased upon TDP43 overexpression in the cell-growth curve analysis. (C) Wound-healing assay in MCF10A cells stably overexpressing Flag-TDP43 or vehicle control. (D) 3D cell culture of MCF10A cells overexpressing Flag-TDP43 or vehicle control for 10 d. (E) Transwell migration assays in MCF10A cells expressing Flag-TDP43 or vehicle vectors. Data are shown as averages ± SEM for three independent measurements. *P < 0.05, ***P < 0.001.
Extensive Splicing Regulation by TDP43 in TNBC Cells.
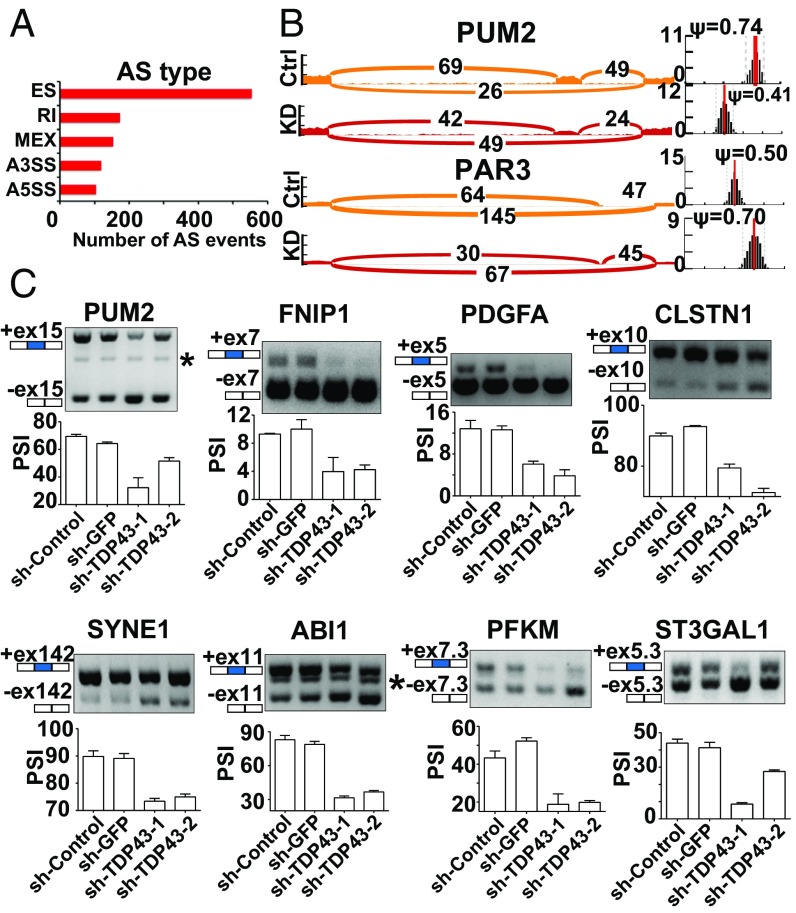
To elucidate the mechanism underlying the pro-oncogenic role of TDP43 in TNBC, RNA-sequencing (RNA-seq) was performed to identify the differential AS events upon TDP43 knockdown by shRNAs in the TNBC cell line MDA-MB231. Using the MISO package, we found that 1,103 splicing events demonstrated significantly changed PSI values (Bayes factor >6 and |ΔPSI| >0.2) after knockdown of TDP43 (Fig. 4A). Gene ontology (GO) analysis revealed that the TDP43-regulated AS targets were enriched in neuron migration, cell–cell adhesion, and regulation of growth, suggesting that TDP43 may affect TNBC progression through AS regulation (Fig. S2G). As shown in Fig. 4B, the PSI values of the two examples changed considerably. The AS changes were also validated by semiquantitative RT-PCR (Fig. 4C). Moreover, we analyzed gene-level expression upon loss of TDP43. The differentially expressed genes (log2 fold change >2 or <−2 and FDR correction, P < 0.05) were clustered in the cell proliferation, endothelial cell apoptotic process, and regulation of TGFβ1 production by GO term analysis (Fig. S2H). These data indicate that TDP43 regulates extensive AS events in TNBC.
Fig. 4.
Extensive splicing regulation by sh-TDP43 in TNBC cells. (A) Quantification of different AS events affected by sh-TDP43: ES, retained intron (RI), alternate promoters (A5SS), alternate terminators (A3SS), and mutually exclusive exons (MEX). (B) Examples of alternative exons affected by sh-TDP43. Numbers indicate read counts. Ψ indicates PSI. (C) Quantitative PCR of different AS events affected by sh-TDP43. Data represent three independent experiments. The asterisk denotes nonspecific amplification.
TDP43 Interacts with SRSF3 in Breast Cancer Cells.
To decipher the TDP43 splicing-regulatory mechanism, we performed immunoprecipitation and mass spectrometry (IP-MS) using antibodies against TDP43 to identify the TDP43-associated proteins in MDA-MB231 cells (Fig. S3A). As shown in Table 1, the IP-MS results included several splicing factors (HNRNPA1, SRSF1, SRSF3, and SRSF7). We then confirmed the interaction of these proteins by coimmunoprecipitation (co-IP) to determine whether these splicing factors interacted strongly with TDP43. The immunoprecipitation assay showed that SRSF3 was pulled down with TDP43 (Fig. 5A), whereas the other proteins (HNRNPA1 and SRSF1) failed to show interaction with TDP43 in either the endogenous or exogenous co-IP experiments using MDA-MB231 cells (Fig. S3 B–E).
Table 1.
Abundant cellular proteins interacting with TDP43 as identified by MS
| No. | Official symbol | Sequence coverage, % | Intensity |
| 1 | HNRNPA1* | 23.4 | 11,001,000 |
| 2 | HIST1H1C | 22.5 | 4,879,400 |
| 3 | SRSF1* | 21.5 | 2,626,400 |
| 4 | TAGLN2 | 17.1 | 19,886,000 |
| 5 | BANF1 | 15.7 | 4,409,500 |
| 6 | SRSF3* | 14.7 | 7,407,700 |
| 7 | SRSF7* | 14.7 | 7,407,700 |
| 8 | MCU | 13 | 4,607,700 |
| 9 | MTRF1 | 12.6 | 193,590,000 |
| 10 | RAI14 | 12 | 29,035,000 |
Proteins related to splicing identified by MS.
Fig. 5.
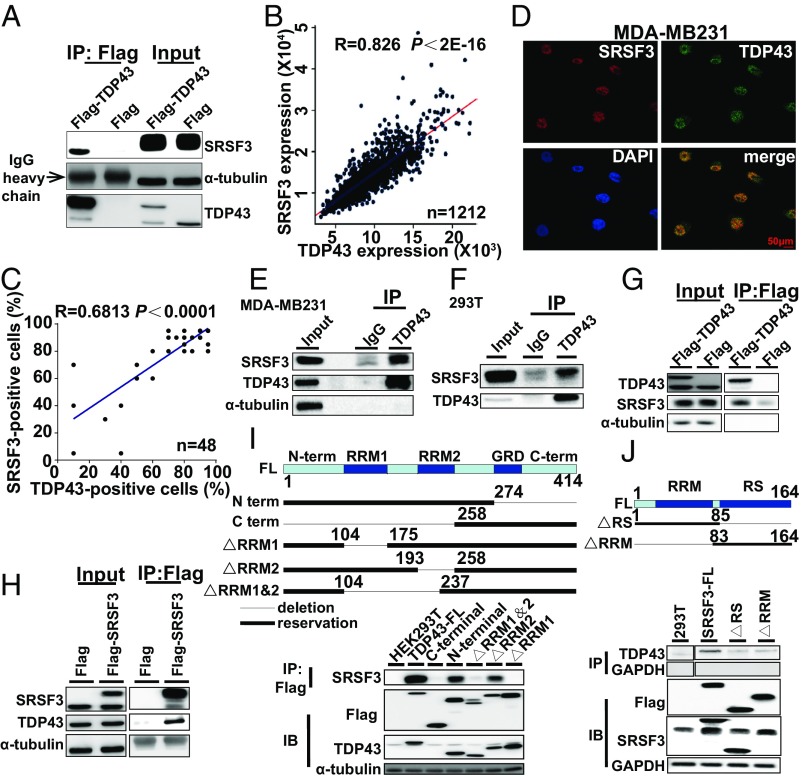
TDP43 interacts with SRSF3 in breast cancer cells. (A) Cellular extracts from MDA-MB231 cells stably expressing Flag (control) or Flag-TDP43 were immunopurified with anti-Flag affinity beads. Immunocomplexes were then immunoblotted using antibodies against the indicated proteins. (B) Analysis of public datasets for expression correlation between TDP43 and SRSF3. (C) Analysis of expression correlation between TDP43 and SRSF3 by immunohistochemical staining of tissue chip data. (D) Subcellular localization of TDP43 (green) and SRSF3 (red) in MDA-MB231 cells determined by immunofluorescence by confocal fluorescence microscopy. (E and F) Endogenous immunopurification with TDP43 antibody in MDA-MB231 (E) and 293T (F) cells. (G and H) Flag-TDP43 pulled down with SRSF3 (G) or Flag-SRSF3 pulled down with TDP43 in HEK293T cells (H). (I and J, Upper) Structures of TDP43 (I) and SRFS3 (J) and their mutants used in immunoprecipitation pull-down assays. TDP43 protein (I) exhibits an N-terminal domain (N-term), two RRMs (RRM1 and RRM2), a glycine-rich domain (GRD), and a C-terminal domain (C-term); whereas SRSF3 full-length protein (J) contains an RRM and an SR-rich domain (RS). (Lower) Flag-mutants of TDP43 pulled down with SRSF3 (I) or Flag-mutants of SRSF3 pulled down with TDP43 (J) from HEK293T cells.
Analysis of expression data from the TCGA database demonstrated a strongly positive correlation between TDP43 and SRSF3 at the mRNA expression level (Fig. 5B), which was also confirmed at the protein level based on the tissue chip staining data (Fig. 5C). We further validated this correlation in various breast cancer cell lines and observed the same positive correlation pattern (Fig. S3 F and G). Double immunofluorescent staining revealed the colocalization of TDP43 and SRSF3 proteins in MDA-MB231 cells (Fig. 5D). The endogenous immunoprecipitation assay also demonstrated that SRSF3 could be pulled down with the TDP43 antibody in MDA-MB231 (Fig. 5E) and 293T (Fig. 5F) cells. Moreover, Flag-TDP43 and Flag-SRSF3 could pull down each other (Fig. 5 G and H), further demonstrating their strong interaction. To determine which domains were responsible for the interaction between TDP43 and SRSF3, various Flag-tagged TDP43- or SRSF3-deletion fragments were pulled down with SRSF3 or TDP43, respectively. The pull-down assay illustrated that only the TDP43 protein containing the RRM1 domain could pull down SRSF3 (Fig. 5I). Conversely, both the N and C terminals of the SRSF3 protein could pull down TDP43, although this interaction was weaker than that with full-length SRSF3 (Fig. 5J).
To test whether the interaction between TDP43 and SRSF3 was RNA-dependent, cell lysis was incubated with DNase and RNase. Results showed that TDP43 could still pull down SRSF3, although the blotting bands in the RNase samples were weaker than those in samples without RNase treatment (Fig. S3H). Moreover, we constructed two point mutants (R171 and R174) and one deletion mutant (TDP43∆106-111), known for their ability, within the RRM1 domain of TDP43, to recognize RNA (9, 34). These constructs all pulled down SRSF3 with positive but weaker bands (Fig. S3I). Overall, TDP43 might interact with SRSF3 directly, and RNA markedly enhanced the affinity of the interaction.
Taken together, our data show that TDP43 interacts with SRSF3, suggesting that these two splicing factors, as important members of the splicing complex, may work together to regulate AS in TNBC cells.
TDP43 Regulates AS in Coordination with SRSF3.
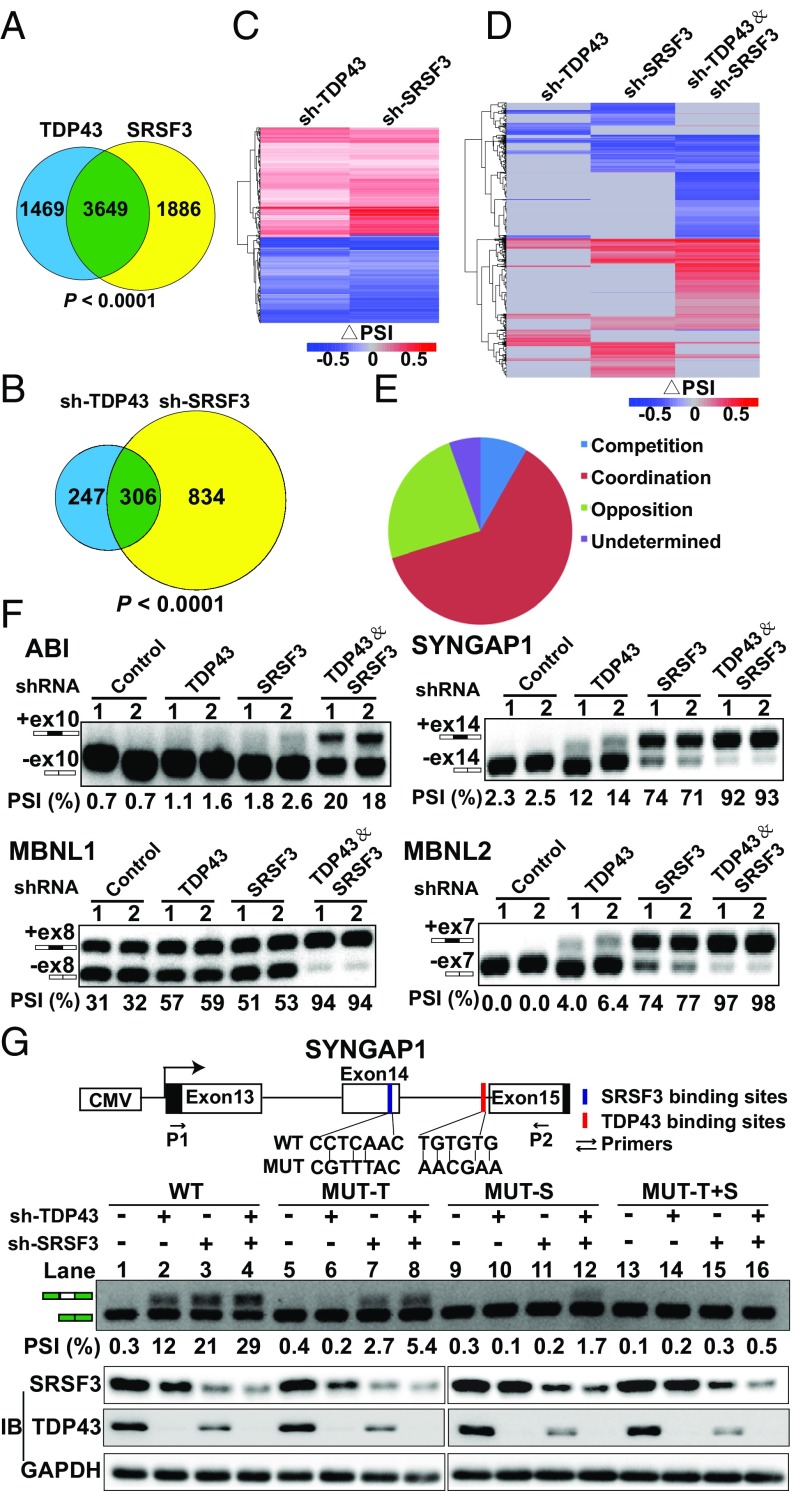
To clarify how TDP43 regulates AS with SRSF3, we assessed whether they possessed direct common binding mRNAs. Both TDP43 and SRSF3 RNA immunoprecipitation sequencing (RIP-seq) were performed to gain insight into their direct downstream mRNAs. Results showed that TDP43 and SRSF3 regulated a large set of overlapping target mRNAs (3,649 mRNAs, more than two-thirds of all binding targets) (Fig. 6A). Changes in AS event values (PSI) were also analyzed based on the RNA-seq data of TDP43 and SRSF3 knockdown in MDA-MB231 cells. We found 553 TDP43-regulated and 1,140 SRSF3-regulated ES-type AS events with significant changes (|ΔPSI| >0.2 and Bayes factor >6). Among them, more than half of the TDP43-regulated AS events (306) were common splicing events, suggesting a large degree of overlap between TDP43-regulated and SRSF3-regulated splicing events (Fig. 6B). Remarkably, most of the 306 common splicing events exhibited the same change in direction, with either positive (red in the heatmap) or negative (blue in the heatmap) values between the TDP43- and SRSF3-knockdown groups (Fig. 6C), indicating that TDP43 and SRSF3 may coregulate the common downstream genes.
Fig. 6.
TDP43 coordinately regulates alternative splicing with SRSF3. (A) Venn diagram showing overlapping mRNAs between TDP43 RIP-seq and SRSF3 RIP-seq binding results. (B) Venn diagram showing coregulated splicing events using RNA-seq data after knockdown of SRSF3 and TDP43 in MDA-MB231 cells. (C) Heatmap of common regulated splicing events after knockdown of SRSF3 and TDP43 in MDA-MB231 cells. (D) Heatmap of all splicing events after knockdown of SRSF3, TDP43, and both in MDA-MB231 cells. (E) Percentage distribution of coordination, competition, opposition, and others based on the heatmap in D. (F) Random examples of coordination splicing events verified by semiquantitative PCR. Two shRNAs (1 and 2) were used for each group. Representative data are from three independent experiments. (G, Top) Minigene assay for verifying coordination between TDP43 and SRSF3. Graphical representation of the control splicing reporter (WT), the MUT-T motif (specifically introducing mutations to TDP43-binding sites) (red), the MUT-S motif (specifically introducing mutations to SRSF3-binding sites) (blue), and the PGZ+T&S motif (introducing mutations to both binding sites). P1 and P2 indicate PCR primers for splicing analysis. (Middle) Semiquantitative RT-PCR analyses were performed for the above four minigenes after TDP43 or SRSF3 knockdown. (Bottom) Western blotting was performed to verify the knockdown of TDP43 and SRSF3.
To further establish the coordination between TDP43 and SRSF3, we performed RNA-seq with both single and double knockdown of TDP43 and SRSF3 in MDA-MB231 cells. In the global splicing-event changes, single TDP43- and SRSF3-knockdown groups generated fewer PSI value changes (events in gray in Fig. 6D), whereas the combination induced a much greater number of PSI changes (events in red or blue in Fig. 6D). After analyzing these PSI values, we found that most events (more than 60%) occurred in coordination (PSI changes of sh-TDP43/sh-SRSF3 were larger than those of sh-TDP43 and sh-SRSF3 individually), whereas 8% of events were in competition (PSI changes of sh-TDP43/sh-SRSF3 were smaller than those of sh-TDP43 or sh-SRSF3), and 24% of events were in opposition (PSI changes of sh-TDP43 and sh-SRSF3 were in the opposite direction) (Fig. 6E). Representative coordination events were randomly selected to confirm the global analysis results. Four selected genes were shown to coordinate (Fig. 6F). These results again suggested that TDP43 coordinated with SRSF3 in the regulation of AS.
To further verify that TDP43 and SRSF3 regulate AS in coordination, SYNGAP1 exon 14 coregulated by TDP43 and SRSF3 (Fig. 6F) was used to perform a minigene reporter assay in MDA-MB231 cells. The wild-type SYNGAP1 minigene construct was created by amplifying the SYNGAP1 gene region from human genomic DNA containing exon 13, intron 13, exon 14, intron 14, and exon 15 (Fig. 6G, Upper). As expected, knockdown of TDP43 or SRSF3 promoted inclusion of SYNGAP1 exon 14 splicing (lanes 2 and 3) in comparison with the sh-control (lane 1). Moreover, the concomitant knockdown by sh-TDP43 and sh-SRSF3 (lane 4) showed a larger increase in exon 14 inclusion relative to sh-TDP43 or sh-SRSF3 alone, consistent with our previous results (Fig. 6G). We thereafter predicted TDP43- and SRSF3-binding motifs using the RBPmap website and identified putative target sequences for SRSF3 and TDP43 located in the SYNGAP1 exon 14 (blue in Fig. 6G, Upper) and intron 14 (red in Fig. 6G, Upper), respectively. This indicated that TDP43 and SRSF3 might control the splicing of SYNGAP1 exon 14 by interacting with their own binding sites. We next introduced mutations in the TDP43- and SRSF3-binding sites (MUT-T and MUT-S, respectively) (Fig. 6G, Lower). As expected, mutation of the TDP43 binding site completely abolished splicing regulation through TDP43 (lane 6). Intriguingly, the increase in exon 14 inclusion by depletion of SRSF3 (lane 7) was also partially abated in comparison with the sh-control (lane 5) when the mutations were introduced into the TDP43-binding sites. Similarly, TDP43-mediated regulation of SYNGAP1 exon 14 splicing was markedly impaired when the SRSF3-binding sites were mutated (lane 10). These results suggested that TDP43 acts in conjunction with SRSF3 to regulate AS and that loss of either protein results in splicing dysregulation in terms of SYNGAP1 exon 14 splicing in MDA-MB231 cells. Collectively, our results suggest that TDP43 collaborates with SRSF3 in the regulation of AS.
Knockdown of SRSF3 Inhibits Breast Cancer Progression.
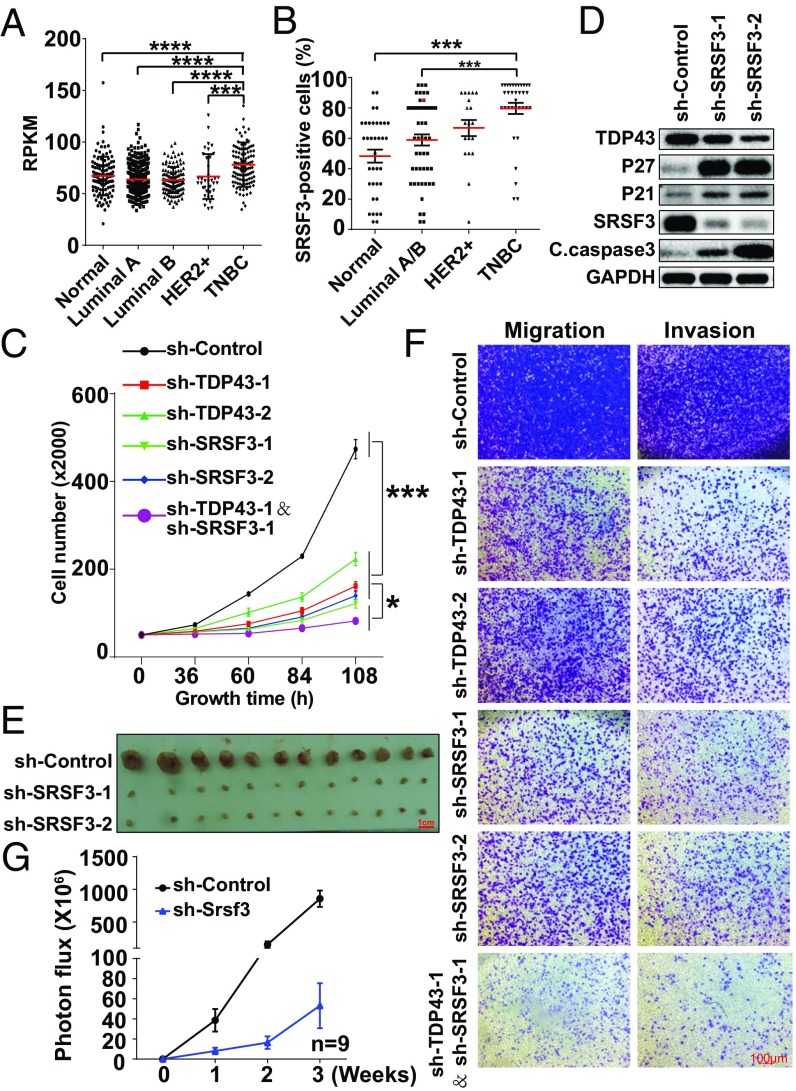
To measure the clinical relevance of SRSF3, we found that SRSF3 mRNA expression was higher in TNBC than in normal tissue and other breast cancer subtypes (Fig. 7A). This high expression in TNBC relative to normal tissue and other breast cancer subtypes was also observed in immunohistochemical tissue chips (Fig. 7B and Fig. S5A), as is consistent with the cultured cancer cell line findings (Fig. S5B). To determine the role of SRSF3 in tumor progression, we investigated cell proliferation after disruption of SRSF3 by shRNA lentivirus (knockdown efficiency is shown in Fig. S5C). The cell-growth curve revealed that knockdown of SRSF3 in MDA-MB231 cells reduced the cell number in a time-dependent manner compared with the control group (Fig. 7C) and HCC1806 cells (Fig. S5 D and E). The BrdU assay showed a dramatic decrease in BrdU-positive signals in MDA-MB231 cells expressing sh-TDP43 or sh-SRSF3 relative to the sh-control (Fig. S5F). These results were further verified and extended by propidium iodide (PI) cell-cycle (Fig. S5G) and annexin V cell-apoptosis analyses (Fig. S5 H and I). Immunoblot analysis also showed a significant increase in cell-cycle–inhibiting proteins (P21 and P27) and apoptotic markers (cleaved caspase-3) following SRSF3 reduction (Fig. 7D). To test whether reduced SRSF3 also negatively affected tumor growth in vivo, a xenograft assay was performed after cells stably expressed sh-SRSF3 or sh-control. A significant decrease in tumor formation was observed upon SRSF3 knockdown relative to the control group (Fig. 7E). The Transwell assay demonstrated that the migration and invasion of cells expressing sh-SRSF3 were markedly suppressed (Fig. 7F). We also estimated cell metastasis by the injection of 4T1 cells expressing sh-Srsf3 into the tail veins of BALB/C mice. Statistical results showed that mice with tail-vein injection of Srsf3-knockdown cells developed fewer lung metastases (Fig. 7G and Fig. S5J). These data suggested that knockdown of SRSF3 inhibited the progression of TNBC in vitro and in vivo.
Fig. 7.
Knockdown of SRSF3 inhibits TNBC progression. (A) Transcript expression levels of SRSF3 in normal tissue and different breast cancer subtypes in the TCGA. (B) Quantification of the percentage of SRSF3-positive staining in breast cancer subtypes by immunohistochemical staining in tissue chips. (C) Cell-growth inhibition upon TDP43, SRSF3, or TDP43 plus SRSF3 knockdown by shRNAs. (D) Western blotting of cell-cycle and apoptosis-related proteins. (E) Tumor size after 2 mo of mammary fat pad growth in the xenograft assay. Each experimental group contained six mice. (F) Cell migration and invasion levels by Transwell assay. (G) Statistical analysis of fluorescence density of metastasis ability following tail-veil injection of 4T1 cells expressing luciferase (n = 9). Data are shown as averages ± SEM for three independent measurements, except in A. ***P < 0.001. All data shown represent MDA-MB231 cells, except where labeled.
To further clarify the coordination relationship of TDP43 and SRSF3 in the control of tumor progression, we employed the concomitant knockdown of TDP43 and SRSF3 in MDA-MB231 cells (Fig. S5C). A more noticeable reduction in cell growth was observed upon the down-regulation of both TDP43 and SRSF3 than with the down-regulation of either TDP43 or SRSF3 alone (Fig. 7C). Moreover, cell migration and invasion ability also declined upon knockdown of TDP43 coupled with SRSF3, relative to knockdown of either alone (Fig. 7F). Thus, both TDP43 and SRSF3 are necessary for cancer progression and work coordinately in TNBC.
Antimetastasis Effects of Reduced TDP43 and SRSF3 Are Mediated by the Splicing Control of PAR3 Exon 12.
To identify the downstream events regulated by TDP43, we first listed the 553 TDP43-regulated ES events upon TDP43 knockdown in MDA-MB231 cells according to our RNA-seq data (Fig. 4A). The list was then narrowed by overlapping well-recognized splicing events in breast cancer compared with normal tissue (27), and we identified 38 high-priority candidate events (Dataset S3). These 38 candidate events were thereafter filtered to identify splicing events in genes reported to serve as important regulators of cancer progression based on previous publications. Eleven candidate events were then selected for qRT-PCR validation upon knockdown of TDP43 and SRSF3 (Fig. S4E). Among them, PAR3 has been reported as a cell polarity protein, and its loss promotes breast tumorigenesis and metastasis (35, 36). NUMB exon 12 splicing is also a frequent tumor-associated AS change observed in lung cancer (37) and acts as a key target of many splicing factors in the regulation of cancer cell proliferation (38). Consequently, we focused on the roles of PAR3 and NUMB in TDP43- and SRSF3-dependent cancer progression.
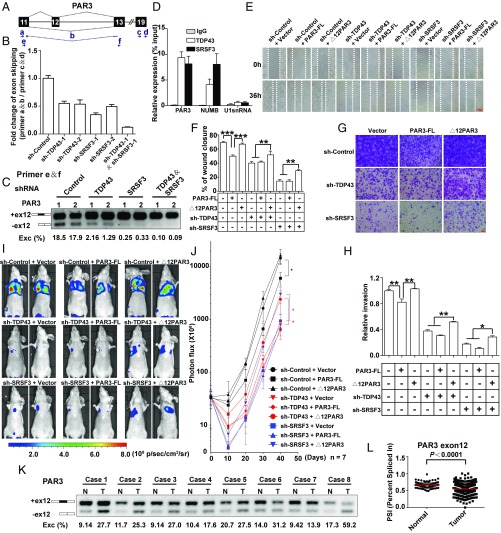
We first confirmed that the exclusion of PAR3 exon 12 was reduced after disrupting the expression levels of TDP43 or SRSF3 using two pairs of primers (Fig. 8 A–C and Fig. S6A). Double knockdown of TDP43 and SRSF3 enlarged the reduction of exon 12 skipping relative to the knockdown of TDP43 or SRSF3 alone, supporting the notion that TDP43 coordinately regulates AS with SRSF3. The RIP assay demonstrated enrichment of PAR3 mRNA with the TDP43 and SRSF3 proteins (Fig. 8D and Fig. S6B), further supporting the possibility of the splicing regulation of PAR3 exon 12 by TDP43 and SRSF3.
Fig. 8.
Antimetastasis effects of reduced TDP43 and SRSF3 are mediated by the control of PAR3 exon 12 splicing. (A) Schematic representation of PAR3 exon 12 splicing. Arrows indicate primers. Primer b spans exon 11 and exon 13. (B) qPCR analysis of changes in PAR3 exon 12 skipping with knockdown of TDP43, SRSF3, or TDP43/SRSF3. (C) Semiquantitative RT-PCR of PAR3 isoforms in the knockdown of TDP43, SRSF3, or TDP43/SRSF3 by shRNA and their control in MDA-MB231 cells. Two shRNAs (1 and 2) were used for each group. (D) Binding of PAR3, NUMB, and U1snRNA mRNA to TDP43 or SRSF3 proteins detected by RIP assay in MDA-MB231 cells. (E and F) Wound-healing assays (E) and statistical analysis (F) in MDA-MB231 cells. Control and TDP43- or SRSF3-knockdown groups were transfected with control plasmids, PAR3-FL, or ∆12PAR3 constructs. (G and H) Transwell invasion assays (G) and statistical analysis (H) in MDA-MB231 cells expressing the indicated vectors. Data shown in B–F represent three independent experiments. (I and J) Luciferase signal intensities (I) and statistical analysis (J) of mice after tail-vein injection with MDA-MB231 cells expressing the indicated constructs. Each experimental group contained seven mice. (K) Splicing of PAR3 exon 12 in paired breast tumor (T) and adjacent normal (N) tissues. (L) PSIs of PAR3 exon 12 in normal (n = 113) and breast cancer (n = 1,088) tissue in TCGA. *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA.
As PAR3 has been reported to regulate cell polarity and migration (35, 36), we supposed that the two PAR3 isoforms caused by PAR3 exon 12 inclusion and exclusion might have different effects on cancer cell migration. To examine the role of PAR3 splicing on TDP43- and SRSF3-dependent changes in cell migration, we generated two constructs by inserting full-length PAR3 (PAR3-FL) and deletion of exon 12 (∆12PAR3) cDNA, respectively (Fig. S6C). As decreased TDP43 or SRSF3 resulted in a splicing shift from ∆12PAR3 to PAR3-FL, we inferred that overexpression of ∆12PAR3 might rescue the changes caused by knockdown of TDP43 or SRSF3. As expected, the wound-healing (Fig. 8 E and F) and Transwell assays (Fig. 8 G and H) showed that overexpression of ∆12PAR3, but not PAR3-FL, could increase migration and invasion in cells expressing sh-TDP43 or sh-SRSF3 relative to those expressing sh-TDP43 or sh-SRSF3 alone. In contrast, significant decreases in cell migration and invasion were also observed upon overexpression of PAR3-FL compared with the control group, partially mimicking the effect seen in TDP43- or SRSF3-knockdown cells. These data revealed that ∆12PAR3 and PAR3-FL had distinct and opposite effects on cell migration and invasion. Reduced TDP43 and SRSF3 promoted the splicing switch from ∆12PAR3 to PAR3-FL, thereby negatively affecting cell migration and invasion.
We also confirmed these results in vivo using tail-vein injection assays. Knockdown of TDP43 or SRSF3 decreased lung colonization ability in MDA-MB231 cells, consistent with our findings in mouse 4T1 cells. Notably, the decrease of signal intensities was partially abolished when coexpressed with the ∆12PAR3 transcript rather than PAR3-FL (Fig. 8 I and J). In addition to migration/invasion, we also examined cell proliferation. Results showed that overexpression of ∆12PAR3 did not affect the phenotypes of sh-TDP43– or sh-SRSF3–driven cell growth (Fig. S6D), indicating that a specific cell-migration change was induced by this splicing switch. To examine whether these two isoforms of PAR3 led to different proteins, MS analysis was performed in MDA-MB231 cells. We successfully identified two peptides, ELNAEPSQMQIPK and ELKAEDEDIVLTPDGTR, which represented exon 12 inclusion and exclusion, respectively (Fig. S6E). These isoforms of PAR3 exon 12 inclusion and exclusion could therefore endogenously encode different proteins.
We also evaluated the clinical relevance of PAR3 exon 12 splicing. Paired breast cancer and adjacent normal tissue samples were surgically obtained from eight human patients. The semiquantitative RT-PCR results revealed that, in comparison with the noncancerous tissue, the exclusion of PAR3 exon 12 (the ratio of the PAR3 isoforms excluding exon 12/total transcript band intensity) increased remarkably in all breast cancer tissues (Fig. 8K), indicating that the ∆12PAR3 might act as an oncogenic isoform. Expression analysis of the TCGA data also found lower PSI values (the ratio of the PAR3 isoforms including exon 12/total transcript) for PAR3 exon 12 in tumor tissues than in normal tissues (Fig. 8L), strongly highlighting this splicing outcome as a potential clinical application. Taken together, these data indicate that depletion of TDP43 or SRSF3 suppresses cancer metastasis through the splicing switch from the oncogenic isoform (∆12PAR3) to the antioncogenic isoform (PAR3-FL).
Antiproliferation Effects of Reduced TDP43 and SRSF3 Are Mediated by the Splicing Control of NUMB Exon 12.
We analyzed the NUMB gene based on our transcriptome splicing analysis and validated changes in NUMB exon 12 skipping using two pairs of primers (38), which revealed the transcript expressions of exon 12 inclusion and exclusion, respectively. Results showed that knockdown of TDP43 or SRSF3 increased exon skipping of the NUMB exon 12, thereby promoting the generation of the NUMB lacking exon 12 form (∆12NUMB) (Fig. S7A). The RIP results revealed an enrichment of NUMB mRNA with both TDP43 and SRSF3 proteins (Fig. 8D). In MDA-MB231 cells with intact TDP43 or SRSF3, overexpression of ∆12NUMB led to significant cell-proliferation inhibition, whereas overexpression of full-length NUMB (NUMB-FL) promoted cell proliferation (black curves in Fig. S7B). Ectopic expression of NUMB-FL, but not ∆12NUMB, increased cell growth when coexpressed with shTDP43 or shSRSF3 (blue and red curves in Fig. S7B), consistent with previous studies showing that NUMB-FL, but not ∆12NUMB, can rescue the inhibition of colony formation caused by the knockdown of splicing factor RBM5 or RBM6 (38). Colony-formation assays also showed that cells expressing NUMB-FL together with sh-TDP43 or sh-SRSF3 grew faster than those expressing sh-TDP43 or sh-SRSF3 alone (Fig. S7C). We also observed higher transcript levels of NUMB with exon 12 in breast tumor samples than in normal samples (Fig. S7D), further confirming the necessity of NUMB exon 12 for tumor progression.
Taken together, our results show that depletion of TDP43 and SRSF3 suppresses breast cancer progression partially through PAR3 and NUMB splicing in TNBC.
Discussion
We uncovered a unique splicing pattern in TNBC relative to normal breast tissue and other breast cancer subtypes. TDP43, as an important member of this splicing factor complex, was necessary for TNBC progression, which was verified by transcriptome analysis and loss/gain-of-function assays. Mechanistically, TDP43 formed a complex and collaborated with SRSF3 in the regulation of AS in TNBC. We also identified a splicing switch from the oncogenic ∆12PAR3 isoform to the PAR3-FL isoform as a downstream splicing event coregulated by TDP43 and SRSF3 (Fig. S7E). Remarkably, these results highlighted TDP43 and ∆12PAR3 as targets for the diagnosis and treatment of TNBC.
In the last few years, TDP43 has been defined as a central pathological protein in neurodegenerative diseases (39). However, studies examining the role of TDP43 in cancer have been less frequently reported, and their conclusions are often contradictory. Some studies have demonstrated that TDP43 is highly expressed in melanoma (40) and hepatocellular carcinoma (41), whereas other research has found that high levels of TDP43 and TRIM16 are good prognostic indicators in neuroblastoma and breast cancer (42). In the current study, we identified TDP43 as a proto-oncogene in TNBC based on a large quantity of clinical data, the TCGA database, and functional assays (Figs. 1–3). Recently, TDP43, interacting with BRCA1, was found to participate in transcription-associated DNA damage in motor neuron cells (43). Because of the high frequency of BRCA1 loss in TNBC (5), it can be inferred that extra TDP43 protein would disassociate the TDP43–BRCA1 complex to undergo pro-oncogenic functions in TNBC. These results imply that TDP43 could be a drug target in breast cancer, especially in TNBC. Interestingly, it has been reported that many antitumor drugs can alter TDP43 expression in cancer cells (44–46), further suggesting that TDP43 might provide a molecular basis for the treatment of breast cancer.
To date, SRSF3 has been identified as a proto-oncogene in several types of tumor. In breast cancer, SRSF3 can regulate HER2 mRNA splicing in HER2+ breast cancer cells, in which the knockdown of SRSF3 leads to a switch from the oncogenic Δ16HER2 to the p100 isoform, which functions as an inhibitor of HER2 signaling (21). In addition, the TCGA database showed that the expression of SRSF3 is higher in TNBC than in HER2+ breast cancer, suggesting that SRSF3 might perform a potential role in TNBC independent of HER2 splicing regulation. Recently, researchers performed SURVIV, a statistical method that outperforms conventional Cox regression survival analysis, to identify alternatively spliced exons correlated with patient survival in large-scale cancer RNA-seq datasets (47). A total of 229 ES events were significantly associated with survival in 682 invasive ductal carcinoma patients from a TCGA breast cancer cohort. They examined the correlation between splicing factors and exon-inclusion levels of survival-associated AS events and found that SRSF3 was significantly linked to 23% (53 of 229) of survival-associated AS events. Furthermore, the higher expression of SRSF3 was associated with shorter survival time and poorer prognosis. These results are consistent with our findings that SRSF3 functions as a pro-oncogene in TNBC progression.
GO analysis of the differentially expressed genes showed that depletion of SRSF3 regulated many cellular biological processes, including cell apoptosis, extracellular matrix organization, and cell adhesion (Fig. S4A). SRSF3-regulated AS events were also identified from our RNA-seq data upon knockdown of SRSF3 in MDA-MB231 cells. GO analysis revealed that the SRSF3-regulated AS targets were involved in the regulation of transcription, RNA splicing, Wnt signaling pathway, and cell cycle (Fig. S4B). These results suggested the important role of SRSF3 in MDA-MB231 cells. The differentially expressed genes and AS targets upon TDP43/SRSF3 knockdown were identified and subjected to GO analysis, revealing that these genes were enriched in cell adhesion, Wnt signaling pathway, and cell migration (Fig. S4 C and D), thus indicating involvement in cancer progression.
The regulatory mechanism mediated by splicing factors, especially for nonspliceosomal RNA-binding proteins, is a foundational issue in the field of AS. Previous research has focused on single splicing factors to clarify their binding motifs to describe their potential roles in AS (48–51). However, specific AS event outcomes are determined by multiple splicing factors (25). Therefore, recent studies have concentrated on the crosstalk between splicing factors in terms of AS (23, 24, 52–54). Based on genome-wide analysis, cross-linking immunoprecipitation sequencing (CLIP-seq), and exon microarray, two SR family proteins, SRSF1 and SRSF2, have been found to exert cooperative and competitive regulation of AS (23). In addition, the RNA-binding proteins ASD-2 and SUP-12 have been reported to bind to nuc-60 pre-mRNA and cooperatively promote a switch from isoform UNC-60A to UNC-60B in Caenorhabditis elegans (54). Our results indicate that TDP43 and SRSF3 form a splicing-regulatory complex in which they coordinately regulate AS in TNBC.
Splice variants frequently contribute to metastatic progression (55). As a fundamental player in cell polarization (56), PAR3 is a strong inhibitor of metastasis in breast cancer (35, 36), lung cancer (57), and pancreatic cancer (58). However, other reports investigating PAR3 have observed the opposite effect in ovarian and skin tumors (59–61). Our results show that the presence and absence of PAR3 exon 12 induce opposite effects on tumor metastasis. It is possible that AS is tissue specific, which might be one reason for the different effects of PAR3 on different tumor types. It would be helpful, therefore, to determine the predominant AS form in different tumors. However, how PAR3 proteins generated by isoforms of exon 12 inclusion and exclusion exhibit different functions in tumor progression remains to be studied.
Experimental Procedures
Materials and methods used for semiquantitative RT-PCR, immunohistochemistry and immunofluorescence, cell culture, the wound-healing assay, the colony-formation assay, cell migration and invasion assays, the lung metastasis assay, immunoprecipitation, Western blotting, plasmid construction, the xenograft assay, and LC-MS/MS were performed using standard protocols, which are described in detail in SI Experimental Procedures. The bioinformatic analyses, including breast cancer subtype classification, physical interaction networks, RNA-seq analysis, and survival analysis, are available in SI Experimental Procedures. Animal procedures were approved by and performed in compliance with the Institutional Animal Care and Use Committee of the Kunming Institute of Zoology, Chinese Academy of Sciences. The clinical samples collection and experiments of this study were approved by the Ethics Committee of the Third Affiliated Hospital of Kunming Medical University. All participants gave written informed consent.
Supplementary Material
Acknowledgments
We thank Drs. Peng Shi, Ceshi Chen, Yongguang Tao, Jumin Zhou, Hu Zhou, and Jiali Li for constructive suggestions and Dr. Christine Watts for text editing. This work was equally supported by Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB13030400 and National Key Research and Development Program of China Grant 2016YFA0100900 (to B.J.). This study was also supported by National Science Foundation of China Grant 31371502 (to B.J.), Yunnan Applied Basic Research Project 2016FB038 (to H.Z.), and Open Project from the State Key Laboratory of Genetic Resources and Evolution Grant GREKF14-05 (to X.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE98016).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714573115/-/DCSupplemental.
References
- 1.Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Livasy CA, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 4.Denkert C, Liedtke C, Tutt A, von Minckwitz G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet. 2017;389:2430–2442. doi: 10.1016/S0140-6736(16)32454-0. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehmann BD, Pietenpol JA, Tan AR. Triple-negative breast cancer: Molecular subtypes and new targets for therapy. Am Soc Clin Oncol Educ Book. 2015:e31–e39. doi: 10.14694/EdBook_AM.2015.35.e31. [DOI] [PubMed] [Google Scholar]
- 7.D’Ambrogio A, et al. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 2009;37:4116–4126. doi: 10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abhyankar MM, Urekar C, Reddi PP. A novel CpG-free vertebrate insulator silences the testis-specific SP-10 gene in somatic tissues: Role for TDP-43 in insulator function. J Biol Chem. 2007;282:36143–36154. doi: 10.1074/jbc.M705811200. [DOI] [PubMed] [Google Scholar]
- 9.Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 10.Colombrita C, et al. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J Biol Chem. 2012;287:15635–15647. doi: 10.1074/jbc.M111.333450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci USA. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moisse K, et al. Divergent patterns of cytosolic TDP-43 and neuronal progranulin expression following axotomy: Implications for TDP-43 in the physiological response to neuronal injury. Brain Res. 2009;1249:202–211. doi: 10.1016/j.brainres.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 13.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: Emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee EB, Lee VM, Trojanowski JQ. Gains or losses: Molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci. 2011;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tollervey JR, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ajiro M, Jia R, Yang Y, Zhu J, Zheng ZM. A genome landscape of SRSF3-regulated splicing events and gene expression in human osteosarcoma U2OS cells. Nucleic Acids Res. 2016;44:1854–1870. doi: 10.1093/nar/gkv1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kano S, et al. Oxidative stress-inducible truncated serine/arginine-rich splicing factor 3 regulates interleukin-8 production in human colon cancer cells. Am J Physiol Cell Physiol. 2014;306:C250–C262. doi: 10.1152/ajpcell.00091.2013. [DOI] [PubMed] [Google Scholar]
- 18.Peiqi L, et al. Expression of SRSF3 is correlated with carcinogenesis and progression of oral squamous cell carcinoma. Int J Med Sci. 2016;13:533–539. doi: 10.7150/ijms.14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sen S, Langiewicz M, Jumaa H, Webster NJ. Deletion of serine/arginine-rich splicing factor 3 in hepatocytes predisposes to hepatocellular carcinoma in mice. Hepatology. 2015;61:171–183. doi: 10.1002/hep.27380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu S, et al. Regulation of CD44E by DARPP-32-dependent activation of SRp20 splicing factor in gastric tumorigenesis. Oncogene. 2016;35:1847–1856. doi: 10.1038/onc.2015.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautrey H, et al. SRSF3 and hnRNP H1 regulate a splicing hotspot of HER2 in breast cancer cells. RNA Biol. 2015;12:1139–1151. doi: 10.1080/15476286.2015.1076610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandit S, et al. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell. 2013;50:223–235. doi: 10.1016/j.molcel.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huelga SC, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Änkö ML, et al. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan M, et al. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res. 2016;44:D1018–D1022. doi: 10.1093/nar/gkv1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buljan M, et al. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol Cell. 2012;46:871–883. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papasaikas P, Tejedor JR, Vigevani L, Valcárcel J. Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery. Mol Cell. 2015;57:7–22. doi: 10.1016/j.molcel.2014.10.030. [DOI] [PubMed] [Google Scholar]
- 30.Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han H, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barash Y, et al. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 33.Park JW, Jung S, Rouchka EC, Tseng YT, Xing Y. rMAPS: RNA map analysis and plotting server for alternative exon regulation. Nucleic Acids Res. 2016;44:W333–W338. doi: 10.1093/nar/gkw410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukavsky PJ, et al. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat Struct Mol Biol. 2013;20:1443–1449. doi: 10.1038/nsmb.2698. [DOI] [PubMed] [Google Scholar]
- 35.Xue B, Krishnamurthy K, Allred DC, Muthuswamy SK. Loss of Par3 promotes breast cancer metastasis by compromising cell-cell cohesion. Nat Cell Biol. 2013;15:189–200. doi: 10.1038/ncb2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCaffrey LM, Montalbano J, Mihai C, Macara IG. Loss of the Par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer Cell. 2012;22:601–614. doi: 10.1016/j.ccr.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Misquitta-Ali CM, et al. Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Mol Cell Biol. 2011;31:138–150. doi: 10.1128/MCB.00709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bechara EG, Sebestyén E, Bernardis I, Eyras E, Valcárcel J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell. 2013;52:720–733. doi: 10.1016/j.molcel.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 39.Ratti A, Buratti E. Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J Neurochem. 2016;138:95–111. doi: 10.1111/jnc.13625. [DOI] [PubMed] [Google Scholar]
- 40.Zeng Q, et al. Identification of TDP-43 as an oncogene in melanoma and its function during melanoma pathogenesis. Cancer Biol Ther. 2017;18:8–15. doi: 10.1080/15384047.2016.1250984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park YY, et al. Tat-activating regulatory DNA-binding protein regulates glycolysis in hepatocellular carcinoma by regulating the platelet isoform of phosphofructokinase through microRNA 520. Hepatology. 2013;58:182–191. doi: 10.1002/hep.26310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim PY, et al. High TDP43 expression is required for TRIM16-induced inhibition of cancer cell growth and correlated with good prognosis of neuroblastoma and breast cancer patients. Cancer Lett. 2016;374:315–323. doi: 10.1016/j.canlet.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 43.Hill SJ, et al. Two familial ALS proteins function in prevention/repair of transcription-associated DNA damage. Proc Natl Acad Sci USA. 2016;113:E7701–E7709. doi: 10.1073/pnas.1611673113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang HY, Chen SB, Guo DJ, Pan SY, Yu ZL. Proteomic identification of differentially expressed proteins in curcumin-treated MCF-7 cells. Phytomedicine. 2011;18:697–703. doi: 10.1016/j.phymed.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 45.Nan YN, et al. Staurosporine induced apoptosis rapidly downregulates TDP- 43 in glioma cells. Asian Pac J Cancer Prev. 2014;15:3575–3579. doi: 10.7314/apjcp.2014.15.8.3575. [DOI] [PubMed] [Google Scholar]
- 46.Jung EJ, Chung KH, Bae DW, Kim CW. Proteomic analysis of novel targets associated with the enhancement of TrkA-induced SK-N-MC cancer cell death caused by NGF. Exp Mol Med. 2016;48:e235. doi: 10.1038/emm.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen S, Wang Y, Wang C, Wu YN, Xing Y. SURVIV for survival analysis of mRNA isoform variation. Nat Commun. 2016;7:11548. doi: 10.1038/ncomms11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Z, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ule J, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 51.Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, et al. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell. 2014;26:374–389. doi: 10.1016/j.ccr.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiou NT, Shankarling G, Lynch KW. hnRNP L and hnRNP A1 induce extended U1 snRNA interactions with an exon to repress spliceosome assembly. Mol Cell. 2013;49:972–982. doi: 10.1016/j.molcel.2012.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohno G, et al. Muscle-specific splicing factors ASD-2 and SUP-12 cooperatively switch alternative pre-mRNA processing patterns of the ADF/cofilin gene in Caenorhabditis elegans. PLoS Genet. 2012;8:e1002991. doi: 10.1371/journal.pgen.1002991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hagen RM, Ladomery MR. Role of splice variants in the metastatic progression of prostate cancer. Biochem Soc Trans. 2012;40:870–874. doi: 10.1042/BST20120026. [DOI] [PubMed] [Google Scholar]
- 56.Goldstein B, Macara IG. The PAR proteins: Fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonastre E, et al. PARD3 inactivation in lung squamous cell carcinomas impairs STAT3 and promotes malignant invasion. Cancer Res. 2015;75:1287–1297. doi: 10.1158/0008-5472.CAN-14-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo X, et al. Par3 regulates invasion of pancreatic cancer cells via interaction with Tiam1. Clin Exp Med. 2016;16:357–365. doi: 10.1007/s10238-015-0365-2. [DOI] [PubMed] [Google Scholar]
- 59.Wysoczynski M, Liu R, Kucia M, Drukala J, Ratajczak MZ. Thrombin regulates the metastatic potential of human rhabdomyosarcoma cells: Distinct role of PAR1 and PAR3 signaling. Mol Cancer Res. 2010;8:677–690. doi: 10.1158/1541-7786.MCR-10-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakamura H, et al. Expression of Par3 polarity protein correlates with poor prognosis in ovarian cancer. BMC Cancer. 2016;16:897. doi: 10.1186/s12885-016-2929-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iden S, et al. Tumor type-dependent function of the par3 polarity protein in skin tumorigenesis. Cancer Cell. 2012;22:389–403. doi: 10.1016/j.ccr.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 62.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Montojo J, et al. GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aken BL, et al. Ensembl 2017. Nucleic Acids Res. 2017;45:D635–D642. doi: 10.1093/nar/gkw1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 68.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.