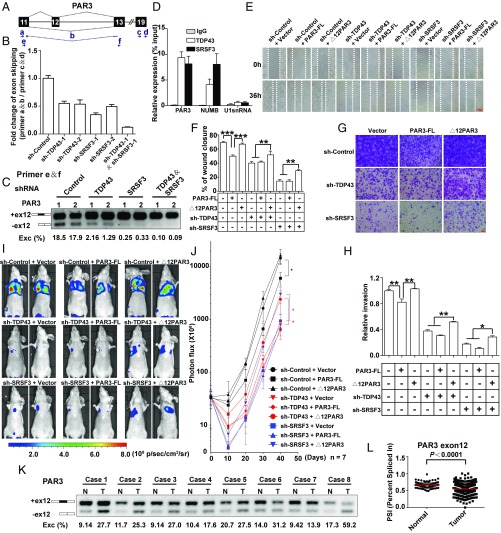
Fig. 8.
Antimetastasis effects of reduced TDP43 and SRSF3 are mediated by the control of PAR3 exon 12 splicing. (A) Schematic representation of PAR3 exon 12 splicing. Arrows indicate primers. Primer b spans exon 11 and exon 13. (B) qPCR analysis of changes in PAR3 exon 12 skipping with knockdown of TDP43, SRSF3, or TDP43/SRSF3. (C) Semiquantitative RT-PCR of PAR3 isoforms in the knockdown of TDP43, SRSF3, or TDP43/SRSF3 by shRNA and their control in MDA-MB231 cells. Two shRNAs (1 and 2) were used for each group. (D) Binding of PAR3, NUMB, and U1snRNA mRNA to TDP43 or SRSF3 proteins detected by RIP assay in MDA-MB231 cells. (E and F) Wound-healing assays (E) and statistical analysis (F) in MDA-MB231 cells. Control and TDP43- or SRSF3-knockdown groups were transfected with control plasmids, PAR3-FL, or ∆12PAR3 constructs. (G and H) Transwell invasion assays (G) and statistical analysis (H) in MDA-MB231 cells expressing the indicated vectors. Data shown in B–F represent three independent experiments. (I and J) Luciferase signal intensities (I) and statistical analysis (J) of mice after tail-vein injection with MDA-MB231 cells expressing the indicated constructs. Each experimental group contained seven mice. (K) Splicing of PAR3 exon 12 in paired breast tumor (T) and adjacent normal (N) tissues. (L) PSIs of PAR3 exon 12 in normal (n = 113) and breast cancer (n = 1,088) tissue in TCGA. *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA.