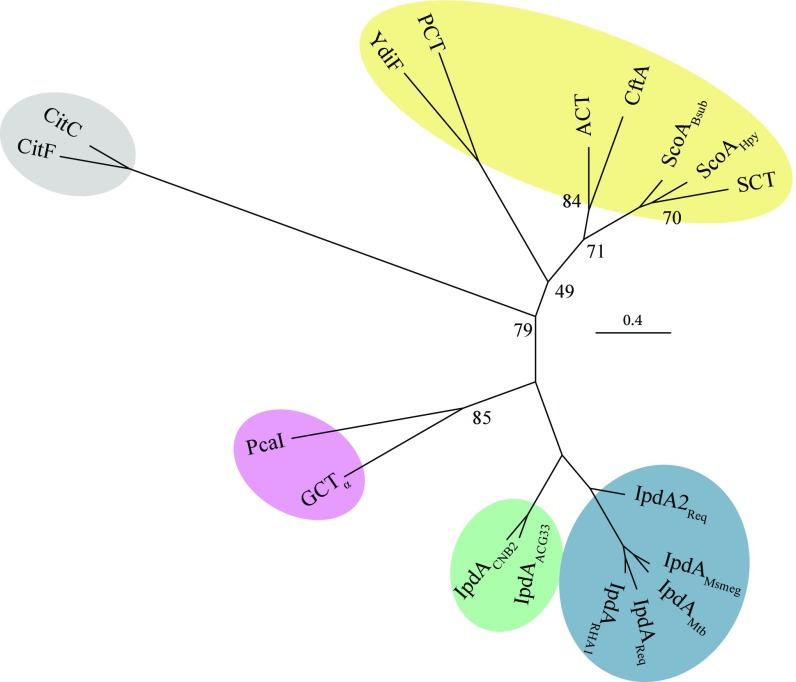
Fig. 2.
Bioinformatic analysis of IpdAB and homologs. Phylogenetic tree displaying IpdA and α-subunits from class I and II CoTs. Shaded regions indicate gram-positive IpdA (blue), gram-negative IpdA (green), or class I β-keto-CoA (purple), class I (yellow) and class II (gray) CoTs. Proteins displayed are IpdA from R. jostii RHA1 (IpdARHA1), R. equi (IpdAReq; IpdA2Req), M. smegmatis (IpdAMsmeg), Mtb (IpdAMtb), Steroidobacter denitrificans (IpdAACG33), and Comamonas testosteroni CNB-2 (IpdACNB2); β-ketoadipyl-CoT from Pseudomonas putida (PcaI); glutaconate CoT from A. fermentans (GCT); citrate lyases from Enterobacter aerogenes (CitC) and Clostridium argentinense (CitF); butyrate-acetoacetate CoT from Clostridium acetobutylicum (CtfA); acetate CoTs from E. coli (ACT and YdiF); succinyl-CoTs from Bacillus subtillus (ScoABsub), Helicobacter pylori (ScoAHpy), and pig heart (SCT); and propionyl-CoT from Clostridium propionicum (PCT). Additional information is available in Supporting Information. Numbers at tree nodes correspond to nonparametric bootstrap values from 100 maximum-likelihood calculations. Bootstrap values >90 are not displayed.