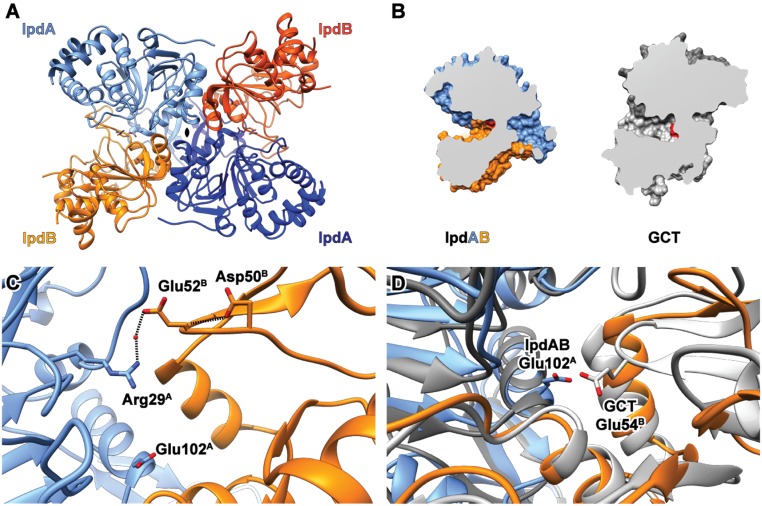
Fig. 4.
Crystallographic structure of IpdABMtb. (A) Heterotetrameric assembly of IpdABMtb. IpdA and IpdB subunits are depicted as blue and orange ribbons, respectively. Active-site residues are shown as ball and stick models. (B) Comparison of substrate binding cleft topology for WT IpdABMtb (blue/orange) and a prototypical class I CoT enzyme GCT (gray; PDB ID code 1POI); catalytic residues are shown in red. (C) Location of potential candidates for the catalytic base in the β-subunit derived from structural alignments with other CoT enzymes are shown in ball and stick. (D) Superposition of the IpdABMtb (blue/orange) and GCT (gray) active sites showing the similar positioning of catalytic residues.