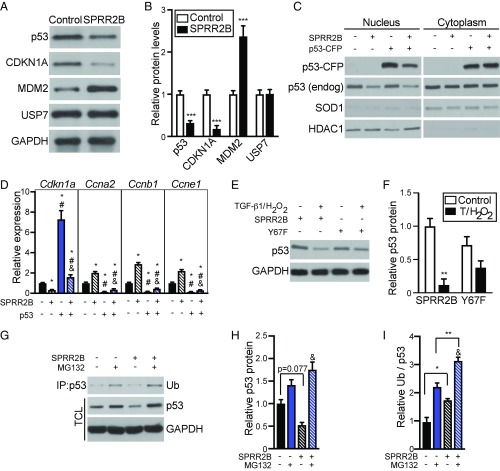
Fig. 6.
SPRR2B facilitates ubiquitin-mediated proteasome degradation of p53. (A) Primary CFs were transfected with Flag-SPRR2B or control and analyzed by Western blot. (B) Quantification of A. n = 3. Analyzed by Student’s two-tailed unpaired t test. (C) Nuclear and cytoplasmic fractions were isolated from NIH 3T3 cells that were transfected with indicated combination of SPRR2B and p53-CFP. Western blot was performed to detect exogenous or endogenous p53. Loading controls are HDAC1 (nucleus) and SOD1 (cytoplasm). (D) qRT-PCR was used to evaluate the expression of genes encoding cell cycle regulators in NIH 3T3 cells that were transfected with p53-CFP with or without SPRR2B (n = 6). qRT-PCR data normalized to Gapdh and analyzed by one-way ANOVA. (E) NIH 3T3 cells were transfected with SPRR2B or SPRR2B-Y67F with or without TGF-β1/H2O2, followed by Western blot for endogenous p53. (F) Quantification of E analyzed by two-way ANOVA with Bonferroni post hoc (n = 3). (G–I) Primary CFs were treated with ZnSO4 (50 μM) and transfected with Flag-SPRR2B with or without MG132 (10 μM). (G) Coimmunoprecipitation of Flag was followed by immunoblot for monoubiquitin (Ub). Western blot was also performed to detect endogenous p53 in total cell lysate (TCL). (H) Quantification of total p53 levels in G relative to GAPDH loading controls. (I) Quantification of immunoprecipitated ubiquitin in G relative to total GAPDH-normalized p53 levels. Analyzed by two-way ANOVA with Bonferroni post hoc (n = 3). All data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001; #P < 0.05 compared with second condition; &P < 0.05 compared with third condition.