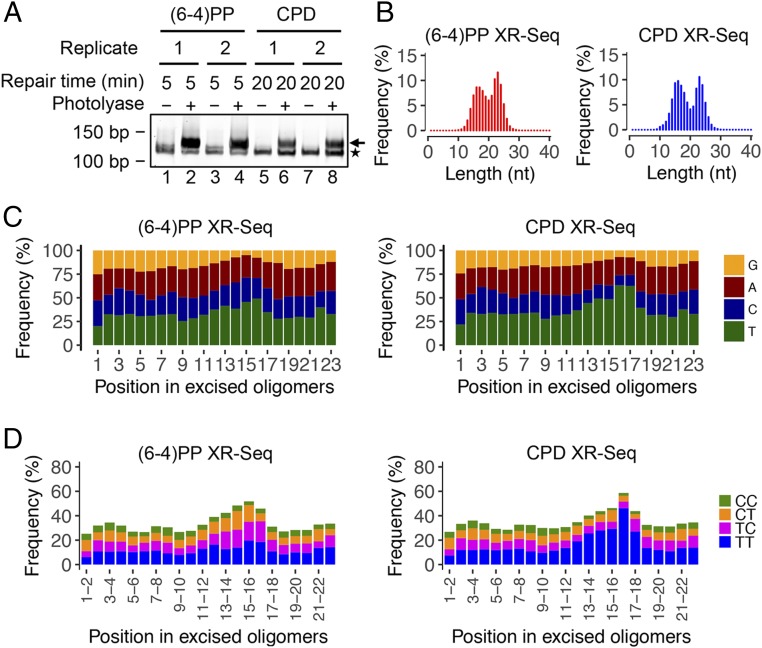
Fig. 3.
Analyses of dsDNA libraries, length distribution, and nucleotide frequencies of the excised oligomers in XR-seq. (A) dsDNA libraries for XR-seq analyzed by 10% native polyacrylamide gel electrophoresis. One percent of ligation products was amplified by 20 cycles of PCR. The arrow on the right indicates the PCR products with inserts. The asterisk on the right indicates adaptor-only PCR products. (B) Length distribution profiles of excised oligomers containing (6-4)PP or CPD from XR-seq. The total read number for each XR-seq was ∼9 million. The repair incubation times for (6-4)PP and CPD XR-seq were 5 and 20 min, respectively. (C) Single-nucleotide frequencies for 23 mers obtained by (6-4)PP and CPD XR-seq. (D) Dipyrimidine frequencies for the 23 mers from (6-4)PP and CPD XR-seq. The enrichment of thymine cytosine (TC) frequency for (6-4)PP XR-seq and that of thymine thymine (TT) frequency for CPD XR-seq are at a position 6 nt from the 3′ end of the excised oligomers.