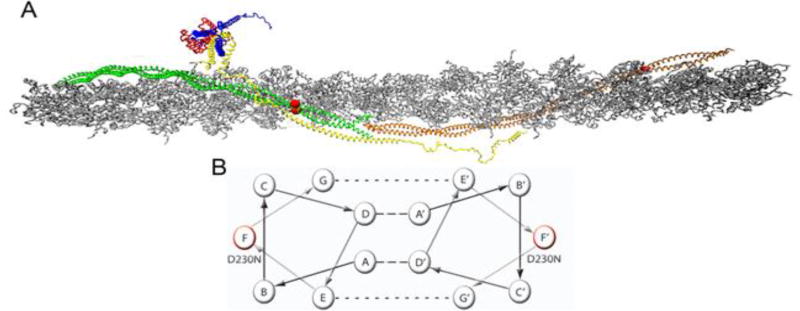
Figure 1.
Atomistic Model of the Cardiac Thin Filament. (A) Average structure taken over 10 ns in a molecular dynamic simulation, (Williams et al. 2016). Grey – filamentous actin (F-actin), Green/Orange – adjacent Tm dimers, Yellow – cardiac Troponin T (cTnT), Red – cardiac Troponin C (cTnC), Blue – cardiac Troponin I (cTnI). Red balls indicate the position of the D230N-Tm mutation. (B) Representative helical wheel of two interacting Tm monomers, monomer one contains a seven heptad repeat (positions A–G) and monomer 2 contains the same repeat (positions A’–G’). Residues in positions A and D are typically hydrophobic (dashed line) and stabilize the core of the dimer. Residues in positions E and G typically salt bridges (dotted line) to further stabilize the dimer. Residues B, C, and F are solvent exposed and can interact with neighboring proteins. The helical position of D230N is marked in red on both monomers.