Abstract
Triple-negative breast cancer (TNBC) exhibits more traits possessed by cancer stem cells (CSC) than other breast cancer subtypes and is more likely to develop brain metastases. TNBC patients usually have shorter survival time after diagnosis of brain metastasis, suggesting an innate ability of TNBC tumor cells in adapting to the brain. In this study, we establish novel animal models to investigate early tumor adaptation in brain metastases by introducing both patient-derived and cell line-derived CSC-enriched brain metastasis tumorsphere cells into mice. We discovered astrocyte-involved tumor activation of protocadherin 7 (PCDH7)-PLCβ-Ca2+-CaMKII/S100A4 signaling as a mediator of brain metastatic tumor outgrowth. We further identified and evaluated the efficacy of a known drug, the selective PLC inhibitor edelfosine, in suppressing the PCDH7 signaling pathway to prohibit brain metastases in the animal models. The results of this study reveal a novel signaling pathway for brain metastases in TNBC and indicate a promising strategy of metastatic breast cancer prevention and treatment by targeting organ-adaptive cancer stem cells.
Keywords: Cancer stem cells, adaptive flexibility, brain metastasis, triple-negative breast cancer, astrocyte-tumor interaction, protocadherin 7, PLCβ-Ca2+-CaMKII/S100A4 signaling
Introduction
Metastatic brain tumors appear in 8–10% of all cancers. Despite advances in neurosurgery and radiotherapy, few patients live longer than a year. Thus, treatment of brain metastases is considered an unmet medical need. While most brain metastases occur at the advanced stages of cancer progression, triple-negative breast cancer (TNBC) usually spreads to the brain rapidly at earlier stages or even before the diagnosis of primary cancer(1). Increased brain metastases at late tumor development stages are primarily attributed to advances in targeted therapy, which improve the management of systemic disease, but the poor bioavailability of these therapies to the brain increases the brain’s potential as a sanctuary site for metastatic disease(2). To this end, targeted therapies with improved brain bioavailability seem promising for disease control(3). However, the treatment for early-developing brain metastases remains a major challenge that limits the patients’ outcome.
About 15 to 25 percent of breast cancer are triple negative, i.e., estrogen receptor-negative, progesterone receptor-negative, and HER2-negative. TNBCs generally do not respond to targeted treatments, so no strategy exists for prevention and control of TNBC brain metastasis. However, brain metastases have consistently been found with increased frequency in younger and premenopausal TNBC patients(4). The shorter time to development of brain metastases in TNBC and shorter survival time after brain metastasis diagnosis(4), may indicate an innate ability of TNBC tumor cells in adapting to the brain.
Cancer stem cells (CSCs), the subpopulation of tumor cells that possess tumor-initiating potential, have been reported to drive tumor metastases(5). The concept of CSCs initiating tumor growth is quite similar to the metastatic process. Although many cells may be shed from the primary tumor, less will survive during circulation to seed a secondary site, and even less will adapt to a new niche and propagate into a clinically apparent tumor(6,7). CSCs possess enhanced plasticity or adaptive flexibility in their cellular microenvironment(8,9). Patients with TNBCs whose tumors have more CSC traits than other breast cancers (8,9) are more likely to develop brain metastases. This led us to hypothesize that brain metastases from TNBC may arise from a brain-tropism CSC population that has enhanced ability to adapt the brain niche.
In our study, patient samples of TNBC brain metastatic tumor and brain-seeking (Br) cell lines were cultured as tumorspheres in neural stem cell conditions to enrich CSC populations. Using sphere-forming assays to assess self-renewal, we showed that brain metastases from TNBC possessed sphere-forming capacity. Intracardiac injection of the patient-derived and Br cell line-derived tumorspheres into immunodeficient mice led to similar multifocal tumor formation and recapitulated tumor cytoarchitecture. Serial injection of patient-derived tumorspheres into mice showed that the putative CSC population can be serially passaged. More importantly, we observed that intracardiac injection of the Br cell line-derived tumorspheres initiated earlier tumor outgrowth in more mice than their corresponding counterpart cell lines, indicating an innate brain-adaptive property of the enriched CSC population.
RNA sequence analysis of the Br cell line-derived tumorspheres vs. their corresponding Br counterpart cell lines identified seventeen differentially expressed genes. Of these, a brain-specific gene protocadherin7 (PCDH7) that normally has very low expression in human breast tissue, had significantly higher expression in the brain metastatic tumors. High PCDH7 mRNA expression was significantly correlated with decreased brain metastasis-free survival in a combined breast cancer patient cohort (n=368). PCDH7 is an important contributor to brain metastasis in a syngeneic lung mouse model(10), although the metastases of lung cancer is quite different from that of breast cancer. We further identified that the increased PCDH7 expression in tumor cells, induced by interacting with astrocytes, preserved the stemness and promoted in vivo tumor colonization through PCDH7-PLCβ-Ca2+-CaMKII/S100A4 signaling. A known drug for selective PLC inhibition, edelfosine, was administered to mouse models to suppress the signaling activation, and the results showed promising efficacy in preventing brain metastatic colonization. These studies demonstrate promise for targeting brain adaptive CSCs to prevent or treat TNBC metastases and indicate a possibility of targeting organ adaptive CSCs to prevent or treat metastasis in general.
Materials and Methods
Cell Lines and compounds
MDA-MB231-Br and CN34-Br human breast cancer brain seeking cell lines were generously provided by Drs. Patricia Steeg and Joan Massague. All other cancer cell lines for studying PCDH7 expression were purchased from ATCC. Normal human astrocytes and human brain microvascular endothelial cells were purchased from Lonza Group Ltd (Allendale, NJ). Cell line characterization or authentication was performed with short-tandem repeat profiling and passaged in our laboratory for less than 6 months after receipt. All cell lines were tested for mycoplasma negative and maintained at 5% CO2 at 37°C. Compound ET-18-OCH3 (edelfosine) was purchased from Sigma Aldrich (St. Louis, MO).
Tumorsphere culture and RNA-seq analysis
Two de-identified TNBC patient brain metastatic tissue specimens were collected in accordance with the Houston Methodist Hospital Institutional Review Board. Written informed consent from the patients were obtained and the studies were conducted in accordance with a recognized ethical guideline Declaration of Helsinki. Samples were mechanically dissociated and subjected to enzymatic digestion with 200µL Liberase Blendzyme (0.2 Wunisch units/mL, Roche) for 15 minutes at 37°C on an incubator rocker (VWR). Undigested tissue was removed, and red blood cells were lysed (RBC Lysis Buffer, Stem Cell Technologies). Cells were washed with phosphate-buffered saline (PBS), subsequently re-suspended in complete NSC (cNSC) media, and plated in an ultra-low attachment plate (Corning). cNSC media is comprised of NSC basal media (1% N2 supplement (Gibco), 0.2% 60µg/mL N-acetylcystine, 2% neural survival factor-1 (Lonza), 1% HEPES, and 6mg/mL glucose in 1:1 Dulbeco’s Modified Eagle Medium and F12 media Gibco), supplemented with 1×antibiotic–antimycotic (Wisent), 20ng/mL human epidermal growth factor (Sigma), 20ng/mL basic fibroblast growth factor (Invitrogen), and 10ng/mL leukemia inhibitory factor (Chemicon). We also used two brain-seeking cell lines cultured in cNSC media: MDA-MB231-Br and CN34-Br cell lines. Cultures were maintained at 37°C, 5% CO2, and media was changed every other day, or as needed.
Total RNA of early-passage tumorspheres derived from MDA-MB231-Br and CN34-Br cell lines were isolated with TRI Reagent (Life Technologies, Carlsbad, CA) and a RiboPure RNA Isolation Kit (Life Technologies) according to the manufacturer’s instructions. rRNA was removed by poly-A selection using oligo-dT beads and mRNA was fragmented and reverse transcribed to yield double-stranded cDNA using random hexamers. cDNA was blunt ended, had an A base added to the 3’-ends, and Illumina sequencing adapters were ligated to the ends. Ligated fragments were amplified for 12 cycles using primers incorporating unique index tags. Fragments were sequenced with an Illumina HiSeq-2000 using single reads extending 50 bases. Raw data were de-multiplexed and aligned to the reference genome using TopHat. Transcript abundance was estimated from the alignment files using Cufflinks. EdgeR was used for differential expression analysis.
Sphere formation assay and limiting dilution analysis
Tumorspheres were dissociated using 5–10µL Liberase Blendzyme in 1mL of PBS for 5 minutes at 37°C. Cells were plated at limiting dilution (1000 to 1 cells per well) in 200µL of cNSC media in quadruplicate in a 96-well plate. After seven days, the number of spheres per well was counted for each dilution, and was used to estimate the mean number of spheres per 2000 cells. For patient samples, this assay estimated secondary sphere formation, whereas cell lines were of passage three or higher. The fraction of negative wells vs. cell dilution was graphed and fitted with a linear regression to estimate stem cell frequency, as in Tropepe et al.(11). Following the assumption that a single stem cell gives rise to one sphere, the proportion of negative wells can be defined by the zero point (F0) of the Poisson distribution: F0 = e−x, where×is the mean number of cells per well. The dilution at which it is expected to have one stem cell (one sphere) per well can be identified by the point at which the line-of-best-fit crosses 0.37 (when×= 1, F0 = e−1 = 0.37)(11).
Microarray data analysis
Two independent datasets of breast tumor cohorts, “EMC-286” (Erasmus Medical Center, n=286; GSE2034) and “MSK-82” (Memorial Sloan-Kettering Cancer Center; n=82; GSE2603), for which microarray and clinical data (metastasis-free survival time) are publicly available were used for the correlation analysis. Researchers defined metastasis-free survival as the “time from randomization to the first evidence of distant metastatic disease, excluding pelvic lymph nodes, or death from any cause”. The classification was implemented in the package “e1071” of R, where the cross-validation for Support Vector Machine (SVM) was set as five-fold and other parameters for the SVM were set as default. We used the receiver operating characteristic (ROC) method to indicate the performances for the patient classification. The area under the ROC curve (AUC) illustrates the accuracy of the classification. Furthermore, we did the brain metastasis-free survival time analysis based on the PCDH7 expression for the breast cancer patients. The survival analyses were implemented in the “survival” package of R. GSE26291 dataset was used to analyze the gene expression of calcium ion channel-related genes, EMT-related genes, cell survival and apoptosis genes, in the MDA-MB231 tumor cells after co-culturing with murine astrocytes for 72 hours, comparing with the tumor cells co-culturing with NIH 3T3 cells.
Oncomine platform data analysis
Relative levels of PCDH7 mRNA expression in normal human tissues were obtained by Oncomine Cancer Microarray database analysis (http://www.oncomine.com). The data was log 2-transformed, with the media set to zero and s.d. set to one. Molecular concept analysis of the differentially expressed 17 genes in tumorspheres was performed using Oncomine platform software.
General animal study procedures
Animal procedures were conducted under the approval of Institutional Animal Care and Use Committee (IACUC) of Houston Methodist Research Institute. For in vivo imaging, all tumor cell lines were stably transduced with Firefly luciferase (F-luc) and EGFP. Female NOD-SCID mice (6–8 weeks-of-age; Charles River Laboratories, Boston, MA) were anesthetized with isoflurane/O2 and injected in the left cardiac ventricle with tumor cells (1.75 ×105 cells in 0.1 mL PBS). Tumor cell growth was monitored by repeated non-invasive bioluminescence imaging twice per week using an IVIS 200 system (Xenogen). “Brain metastasis-free survival” was defined as the interval from tumor cell injection until the emergence of brain metastatic bioluminescent signals in mouse brain. In vivo compound treatment with the PLC selective inhibitor ET-18-OCH3 (ip, once daily, 30 mg/kg/day) started 10 days after tumor cell injection. Mouse physical conditions were monitored once daily and animals were euthanized with CO2 asphyxiation when neurological impairment was evident or physical condition scores were ≤ 2. Whole brains were removed and 10µm brain sections were serially cut. One section for every 100 µm was stained with hematoxylin and eosin (H&E), or anti-CD34, anti-Ki67, anti-pPLCβ and anti-PCDH7 according to standard procedures. Whole-slide montage images were acquired with an Olympus IX81 automatic microscope (HMRI Advanced Cellular and Tissue Microscope Core Facility) and a software algorithm that we developed was used for segmentation and quantification analysis(12).
Intravital two-photon microscope imaging
Considering a closed window will better preserve the physiological microenvironment for tumor cells and cause less damage to mouse for the long-time imaging, we chose to establish the closed thinned-skull window. Surgery was done under aseptic conditions. The mouse’s head was fixed by a stereotactic apparatus. The skin on top of the frontal and parietal regions of the skull was cleaned with antimicrobial betadine solution. A longitudinal incision of the skin was made between the occiput and forehead. The skin was then cut in a circular manner on top of the skull, and the periosteum underneath was scraped off to the temporal crests. A 5×5 mm square was drawn over the frontal and parietal regions of the skull. Using a high-speed air-turbine drill (CH4201S; Champion Dental Products, Placentia, CA) with a burr tip size of 0.5mm in diameter, a groove was made on the margin of the drawn square. This groove was made thinner by cautious and continuous drilling of the groove but not drilling through the skull. Then, the same cautious drilling procedure was applied to thin the skull within the groove area until the skull becomes transparent and the vessels are visible. Cold saline was applied during the drilling process to avoid thermal injury of the cortical regions. A cover-glass was then glued on top of the skull for three purposes, 1) provide rigidity, 2) inhibit bone regrowth, and 3) reduce light scattering from irregularities on the bone surface. Imaging depths of up to 250µm below the cortical surface can be achieved using two-photon laser scanning microscope. The surgery took an hour in each case, the animal was positioned on a heating pad (37°C) during the surgery and until they recovered from the anesthesia. 1 × 105 tumor cells in 20μl PBS were injected through a biocompatible microrenathane catheter (Braintree Scientific Inc., Miami, FL) into the common carotid artery, and a 3mm-length catheter “stent” was secured in place with two sutures to re-establish blood flow to the carotid artery.
Intravital two-photon imaging of tumor-bearing mice was performed with an Olympus FV1000-MPE microscope with a 25x, 1.05 NA water immersion objective with a correction collar. The laser-light source consisted of a standard femtosecond-pulsed laser system (Mai Tai HP with DeepSee, Newport/Spectra-Physics, Irvine, CA) for excitation of fluorophores (740–950 nm). Fluorescent signals were collected via a dichroic mirror (DM670LP) and sent to photomultiplier tube (PMT) detectors behind appropriate filter cubes to quantify GFP and Texas Red. Texas Red dextran (70 kDa, Invitrogen, cat # D1830) was used to identify the vasculature. We initially imaged 10–15 random fields (512 × 512 µm at 512 × 512 pixels) for a depth of 50–250 µm using 5-µm steps beginning at the brain surface, followed by 4–6 cortical regions of interest, where tumor cells were visible, over time (days 1, 3, 6, 10 and 14). If tumor cells disappeared in one area, we imaged this area a final time, and then selected another area of interest and followed. We retrieved the same cells over time using a vascular patterns and unique branch points of the cortical vasculature as landmarks.
Image analysis
Images were reconstructed in 3D and through time using Image J. We quantified 108 individual metastasizing cells in the PCDH7shRNA cell line (N=6 mice) and 102 in the control cell line (N=5 mice) over 14 days. In addition, we imaged cancer cells for 30–60 min directly after intra-arterial injection (PCDH7shRNA: 58 cells, N=4 mice; control: 69 cells, N=4 mice). We measured the same brain regions over time and quantified for each metastatic nodule: from day 1, for each individual cancer cell over time, we noted that a fraction of cancer cells were intravascular, extravasated and perivascular as single cells, and perivascular as multiple cells (≥ 4). Dormant cells were noted during the experiment. Only cancer cells directly contacting a vessel were “perivascular”. Dormancy was defined as lack or arrest of cancer cell proliferation without signs of regression during the duration of one experiment. Micrometastases were defined as multicellular cancer cell clusters of 4–10 cells (generally indicates a metastasis diameter of < 50µm), and macrometastases as clusters of > 10 cells (> 50µm). These are mouse tumor equivalents of an MRI-detectable human brain metastasis (5 mm).
Statistical Tests
Data are expressed as means ± SEM. To compare groups, we used the Student’s two-tailed t test or the ANOVA with post-hoc test (Fig 3C, 4G and 5E). To assess correlations, we calculated the Spearman’s rank correlation coefficient. To compare frequencies of metastasis and metastasis-free survival, we used Fisher’s exact test and log-rank test. P<0.05 was regarded as statistically significant. We performed all calculations with SigmaPlot statistical software (version 11.2; Systat Software Inc. Chicago, IL).
Fig. 3. Functional roles of PCDH7 in brain metastasis.
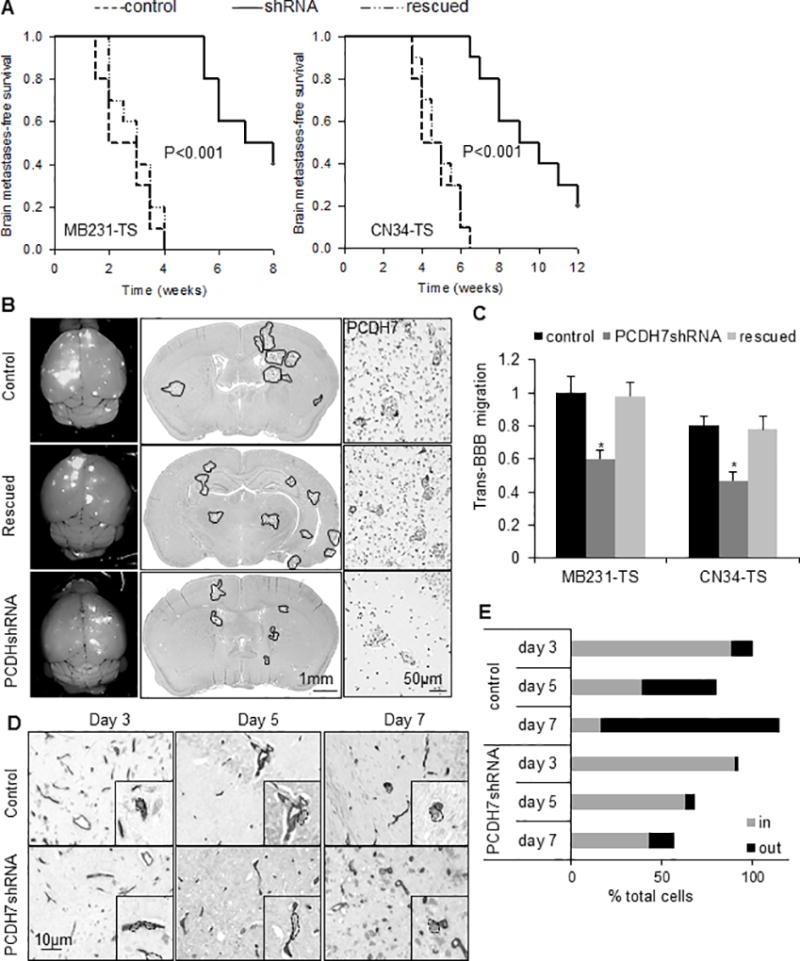
A. Kaplan-Meier curves for brain metastasis-free survival of mice injected with tumorsphere cells derived from MB231-Br (left) or CN34=Br (right) models expressing the shRNA targeting PCDH7, the control shRNA or the PCDH7 rescued. n=10 mice per group. P value was determined using log rank test. B. Representative images showing metastatic lesions in mouse whole brain (left column), H&E stained brain section (middle column), and PCDH7 immuno-reactivities in the brain metastases (right column). It’s noted that high expression of PCDH7 in both tumor cells and surrounding astrocytes in control group was diminished by PCDH7shRNA. n=5 mice per group. C. Transmigration of the indicated cells through the in vitro BBB system. * P<0.05, vs. control shRNA. D. Representative images showing intravascular and extravasated tumor cells in mouse brain sections. MB231-TS cells were visualized on 10µm-thick brain sections by anti-human CD44 and blood vessels by anti-mouse CD34 using immunohistochemistry. Black rectangles are higher magnification single cell images. CD44+ tumor cells were segmented by dash lines. E. Percentage of cancer cells located inside (in) vs. outside (out) blood vessels at indicated days after intracardiac tumor cell injection. Intravascular and extravasated tumor cells were counted in every fourth section throughout the entire mouse brain. Data was relative to “control group day 3”. It’s noted that the % total cells > 100% is due to the cell proliferation, and the % total cells < 100% is due to cell disappear. n=3 mice per each time point.
Fig. 4. PCDH7 mediates the interaction between cancer cells and astrocytes to promote tumor colonization.
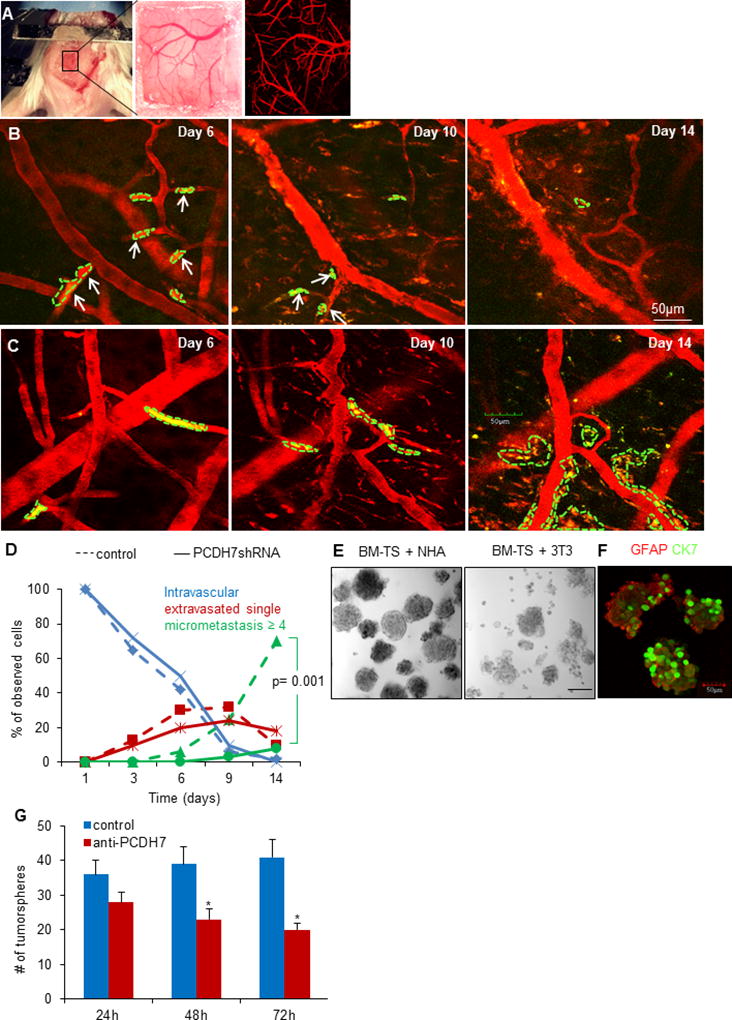
A. A thinned-skull window with perfused brain vessels (Texas-red dextran, red). B. Intravital two-photon microscopy images showing tumor cell arrest and extravasation in mouse brain at day 6, and disappeared at day 10 and day 14 (indicated as solid white arrows) for PCDH7shRNA MB231-TS cells, while the control shRNA cells formed colonization at day 14 (C). It’s noted that the blood vessels became tortuous and leaky after day 6 and remodeled at day 14. D. Extravasation and colonization of the indicated cells in mouse brain over time. n=4 mice per group. E. Representative bright-filed images of tumorspheres at passage 2 when co-culturing BM-TS cells with NHA or normal fibroblasts 3T3 cells. F. Representative immunofluorescent images showing the heteotypic spheres formed by cancer cells (stained for CK7) and astrocytes (stained for GFAP). G. Effects of PCDH7 neutralization by anti-PCDH7 antibody (1:100) on tumorsphere formation (diameter >50µm) in the dissociated BM-TS cells and NHAs co-culturing system. * P<0.05, vs. control.
Fig. 5. Tumor-astrocyte interaction activates PCDH7-PLCβ-Ca2+ signaling to promote colonization.
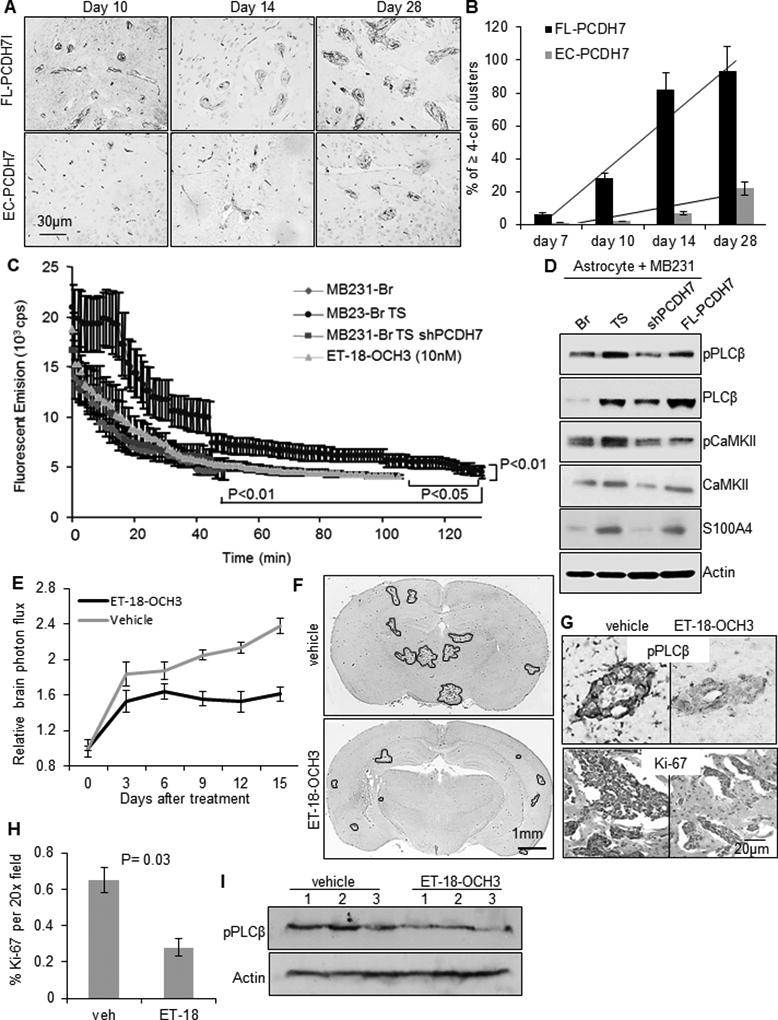
A. Representative immunohistochemistry images of metastatic colonies in mouse brain sections with intra-cardiac injection of indicated cells. Tumor cells were visualized with anti-human CD44 antibody and blood vessels were visualized with anti-mouse CD34 antibody. B. Percent of ≥ 4-cell clusters located outside vessels at indicated days after tumor cell injection (n=3 mice/time point/cell line). C. Baseline level of cytoplasmic Ca2+ in the indicated tumor cells measure by ratiometric emission of calcium-saturated Asante of the tumor cells when co-culturing with astrocytes. D. Signal activity of PLCβ, CaMKII and S100A in the indicated MB231 cells when co-culturing with normal human astrocytes. E. In vivo effects of PLC selective inhibitor ET-18-OCH3 (i.p., 30mg/kg, once daily) on brain metastatic tumor growth. n=10 per group. P=0.04, determined by ANOVA post-hoc test. F. Representative whole-brain sections showing macro-metastatic lesions of ET-18-OCH3 or vehicle-treated mice at the end of the treatment. G. Representative immunostained images of pPLCβ1 and Ki-67 in brain sections. H. Quantification of the % of Ki-67-positive tumor cells in the brain sections. P=0.03, determined by student’s t-test. I. Western blot of pPLCβ1 expression in mouse brain lysates. N=3 per group.
Results
Patient brain metastatic tumor and brain-seeking cell lines form self-renewable tumorspheres
We obtained fresh brain metastases tissue from two TNBC patients and cultured them as primary and then secondary tumorspheres in neural stem cell media. As a sphere is considered to represent all progeny from a single stem cell, tumorsphere formation reflects the presence of a stem cell population(11). We observed that the second sphere-forming efficiencies of the patient tissue were 27 spheres/2000 cells [SD=21] and 11 spheres/2000 cells [SD=3.4] (P=0.39), where secondary sphere formation is a hallmark of the self-renewal property of stem cell(13). Stem cell frequency was estimated via limiting-dilution analysis, and the median frequencies for the two patient samples were 1 sphere/127 cells (range 1/35–1/217 cells) and 1 sphere/200 cells (range 1/125–1/256 cells); they were not statistically different (P=0.40). We supplemented our work with representative human breast cancer Br cell lines. The MDA-MB231-Br and CN34-Br are both derived from brain metastases originating from TNBC tumors(14). Both cell lines formed spheres (MDA-MB231-Br, 153 spheres/2000 cells [SD=94]; CN34-Br, 53 spheres/2000 cells [SD=13]), and their sphere-forming capacity is higher than that of cultured patient samples, which may due to prolonged culture in increasing stem cell frequency(15), as evidenced by frequencies of one sphere/18 cells and one sphere/40 cells for MDA-MB231-Br and CN34-Br cultures, respectively. Despite the increase in sphere-forming capacity, the Br cells lines and patient brain metastases-derived tumorspheres were of similar morphology and size (Fig. 1A).
Fig. 1. Brain metastasis models derived from brain metastasis-derived tumorspheres.
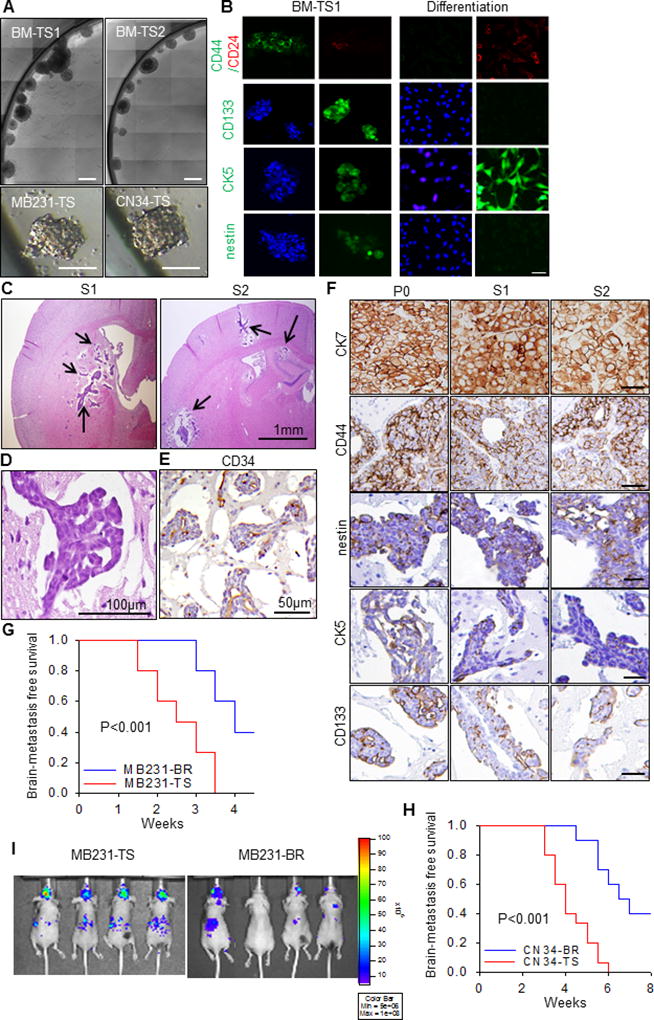
A. Bright-field images of representative tumorspheres cultured from fresh brain metastases tissue from two TNBC patient undergoing neurosurgical resection, i.e., BM-TS1 and BM-TS2, and brain seeking cell lines, i.e., MDA-MB231-TS and CN34-TS. Scale bar is 50µm. B. The BM-TS1 tumorspheres were stained with antibodies to antibodies to CD44, CD24, CD133, CK5 and nestin. For differentiation, BM-TSs were disrupted and the cells plated onto glass coverslips in medium supplemented with 5% fetal calf serum. After 10 days of adherent culture, the cells were stained with the same antibodies. scale bar: 20µm. C. Representative H.E. staining images showing the intraparenchymal multifocal S1 and S2 brain metastatic tumors in mice. Mice were injected with 1.75×105 BM-TS dissociated cells through the left ventricle. D. The cytoarchitecture of the S1/S2 tumor. E. The vessel cooption growth pattern of the S1/S2 tumors. F. The expression pattern of CK7, CD44, nestin, CK5 and CD133 in the original patient P0 brain metastatic tumor and serially passaged S1 and S2 tumors. G–H. Kaplan-Meier survival curves for brain metastasis-free survival of tumorsphere derived models (MB231-TS, CN34-TS) in comparing with brain seeking cell line derived models (MB231-Br, CN34-Br). P values were determined by log rank test. N=10 in each group. I. The representative bioluminescent imaging of the brain metastases in MB231-TS and MB-231-Br models.
Breast tumors from patients with the brain as the first metastatic site were negative for estrogen and progesterone receptor but frequently expressed CK5, nestin, and prominin-1 (CD133)(16). Interestingly, both nestin and CD133 are considered to be CSC markers for glioblastoma(17). Similarly, an in vitro selection of a CSC population from the TNBC cell line identified CD133 and CD44 as marker proteins for these cells(18). We examined the expressions of these markers in the patient–derived spheres (CK5, nestin, CD133, and CD44) (Fig. 1B), correlating with the presence of a stem-like population.
Patient-derived tumorspheres are serially transplantable in mice and form intraparenchymal multifocal tumors
The gold standard for identification of a CSC population is serial tumor formation in immunocompromised mice(19). Since the brain lacks a lymphatic system, cancer cells can only metastasize to the brain via the blood stream(20). We thus performed serial intracardiac injections into NOD-SCID mice using patient-derived tumorspheres to assess their ability to be serially passaged in vivo. Two parental (P0) tumors were cultured into tumorspheres to select for CSCs. Three mice each were injected with 1.75×105 P0 tumor cells (n=6 total). Each P0 derivative from the subsequent (S1) tumors was similarly cultured and injected to give rise to S2 tumors (n=6 total). The two patient P0 tumors derived S1 and S2 tumors in all 12 mice, forming intraparenchymal multifocal masses, and the tumor cells are cohesive, cuboidal or columnar cells arranged in sheets, tubules or acinar structures, and have moderate-to-abundant cytoplasm (Fig. 1C–D). The tumor growth was primarily observed around small blood vessels (co-option) (Fig. 1E). This is consistent with clinical presentation of brain metastases from breast cancer, which are typically multifocal and intracerebral, and less commonly solitary and leptomeningeal. Both S1 and S2 tumors (n=4) were stained with the same markers used to clinically diagnose patient brain metastases, i.e., cytokeratin 7 (CK7), ER, PR, and HER2. The CK7-positive and ER/PR/HER2-negative staining profiles and patterns in the S1/S2 tumors were identical to those of the original patient brain metastasis (Fig. 1F). The expressions of nestin and CD44 were abundant in the S1/S2 tumors, and CD133 and CK5 were moderately expressed (Fig. 1F). These indicate that injection of brain metastasis tumorspheres in mice recapitulate the patient tumors; it also demonstrates that tumorspheres possess the CSC capacity to differentiate into various tumor cell types in vivo.
Brain-seeking cell line-derived spheres exhibit enhanced capacity in inducing brain metastases than their counterpart cell lines
The Br cell lines and the derived tumorspheres have stable luciferase expression, which enables us to concurrently compare their brain metastatic kinetics in vivo. Although brain metastasis occurred in all the mice injected with Br cells or tumorsphere cells (MDA-MB231-Br and CN34-Br) at the same cell number of 1.75×105 or 1.75×104, the tumorsphere cells initiated brain metastases earlier (P<0.001) (Fig. 1G–I). However, our data could not show that the tumorsphere cells are more tumorigenic at lower numbers than their Br counterparts, because injection of low number cells (1.75×103 and 1.75×102) failed to induce brain metastasis in almost all mice with tumorsphere or Br cell injection. It is likely due to the limited tumor cell arrest at blood vessels and the low efficiency of metastatic outgrowth. It has been reported that 8–10% of the total injected tumor cells are able to arrest at the blood vessel branches when injected through carotid artery(7), and the rate is expected to be even lower when injected through left ventricle. Further, >95% of the arrested cells will die during development and only less than 5% cells will continue to grow into macro-metastasis(7,21). Nonetheless, the data showing earlier detection of brain metastases in 100% of mice with the tumorsphere cells compared to regular Br cells suggests that the CSCs have an enhanced adaptation ability to colonize in the brain.
High PCDH7 expression in brain metastases
We performed RNA sequence analysis of the early-passage tumorspheres derived from both MDA-MB231-Br and CN34-Br cell lines. Comparison of the common differentially expressed genes between the two lines identified 16 up-regulated genes in both tumorspheres relative to the Br cells (P<0.01, supplementary Table. 1), and 1 down-regulated gene in both tumorspheres, i.e., STC1. Interestingly, the molecular concept analysis using Oncomine platform software showed that the 16 genes have a strong association with the literature-based concept of molecular signature of human embryonic stem cells (P=7.69E-6, Q=0.01)(22).
Among the 17 genes, PCDH7 is the only one that normally expresses exclusively in human central nervous system (Fig. 2A and S1A) and mouse brain(23). Up-regulated mRNA expression of PCDH7 was reported in both MDA-MB231-Br and CN34-Br cell lines(14). PCDH7 protein was highly expressed in the derived tumorspheres: 5.5-fold higher than MDA-MB231-Br (P=0.004) and 4-fold higher than CN34-Br (P=0.008)(Fig. 2B). PCDH7 is also highly expressed in TNBC patient brain metastasis-derived tumorspheres and moderately expressed in normal human and mouse astrocytes (NHA and NMA), and human brain microvascular endothelial cells (HBVEC)(Fig. 2C). However, other members in the PCDH family are not differentially expressed between tumorsphere and Br cells (Fig. S1B).
Fig. 2. High expression of PCDH7 in brain metastasis.
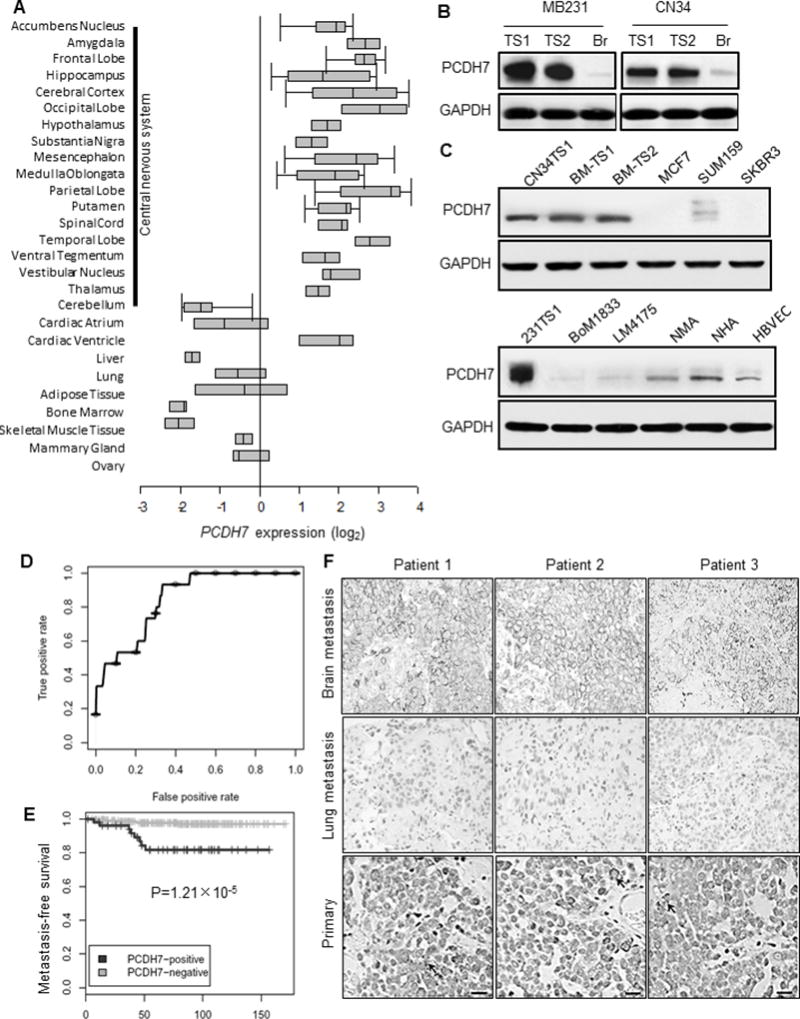
A. Oncomine database analysis of PCDH7 expression in normal human tissue (Neurogenetics 2006 7:67–80). B. Western Blot analysis of PCDH7 in independent brain metastasis-derived tumorspheres (TS1 and TS2) and corresponding brain-seeking (Br) cell lines. C. Protein expression of PCDH7 in TNBC patient brain metastasis tumorspheres (BM-TS1 and 2) and various cell models. MCF7, SUM159, SKBR3: human breast cancer cell lines; BoM1833: MB231 bone-seeking cell line; LM4175: MB231 lung seeking cell line; NMA: primary normal mouse astrocytes; NHA: normal human astrocytes; HBVEC: human brain microvascular endothelial cells. D. Receiver Operating Characteristic (ROC) curve for PCDH7 expression in primary breast tumor samples of brain metastatic patients using the combined 368 microarray data (MSK-82 and EMC-286 cohort). E. Kaplan–Meier curves showing the brain metastasis-free survival of patients with positive or negative PCDH7 expression in the combined cohort of 368 breast cancer patients, P=1.21×10−5 determined by log rank test. F. Representative PCDH7 immunohistochemistry staining of matched patient tissue sections of brain metastasis, lung metastasis and primary breast tumors. Scale bar: 20µm.
To explore the clinical relevance of PCDH7, we retrospectively studied breast cancer patients and noted high PCDH7 mRNA expression significantly correlated with brain relapses (AUC=0.849, Fig. 2D) and decreased brain metastasis-free survival (P=1.21×10−5, Fig. 2E) in a combined breast cancer patient cohort (n=368, EMC-286+MSK-82). PCDH7 expression was not correlated with bone or lung metastasis in these clinical cohorts (Fig. S1C–D). We further measured PCDH7 protein in 34 primary breast tumors and 29 brain metastatic tumors of TNBC patients (4 are matched samples with both primary and brain tumors) (Fig. 2F). Most of the brain metastatic samples (27/29) had strong PCDH7 staining (H score 3+, see Methods), including 4 samples with matched primary tumors, and the 2 remaining samples had intermediate immunoreactivity (H score 2+). Three of the 4 matched primary tumors had sporadic staining for PCDH7, whereas 91% (31/34) of primary breast cancer samples were negative for PCDH7 (H score 0 and 1+; Supplementary Table. 2). In addition, PCDH7-positive staining was not observed in the nine lung metastatic tissue samples (Fig. 2F). Thus, PCDH7 may be a key contributor to TNBC brain metastases.
Functional role of PCDH7 in brain metastasis
To determine the functional involvement of PCDH7 in brain metastasis, knockdown of PCDH7 by shRNAs and rescue of PCDH7 gene expression in brain metastatic tumorspheres were performed (Fig. S2A–B). Tumorsphere cells (1.75 × 105) with knockdown PCDH7 (PCDH7 shRNA), rescued PCDH7, or control shRNA were injected into the left ventricle of mice. In the MDA-MB231-Br tumorsphere model, three weeks after cell inoculation, 70% of mice injected with control shRNA cells had detectable brain metastases, but no PCDH7 shRNA mice did (P<0.001; Fig. 3A-left). Then, five weeks after inoculation, all controls had brain metastases, but only 4 in the PCDH7 shRNA group developed brain metastases by 6 weeks post inoculation. 40% of the mice in the PCDH7 shRNA group had no brain metastasis as confirmed by histology until the end of 8 weeks. Furthermore, knockdown of PCDH7 decreased metastatic lesion numbers and areas in mouse brain sections (P<0.05, Fig. 3B and Supplementary Table. 3). Delay and prevention of brain metastases by PCDH7 knockdown occurred similarly in the CN34-Br tumorsphere mouse model (Fig. 3A-right). Mice injected with PCDH7-rescued cells had brain metastases similar to controls (Fig. 3A–B). Knocking down PCDH7 did not modify mammary tumor growth and lung colonization in vivo (Fig. S2C–D). These findings further confirm that PCDH7 is a key player in brain metastatic formation.
We then evaluated the role of PCDH7 in specific brain metastatic steps. We found that knockdown of PCDH7 decreased tumor cell extravasation through an in vitro blood-brain barrier system and in mouse brain (Fig. 3C–E). Furthermore, as examined by intravital two-photon microscope, the PCDH7 shRNA cells showed a significant defect in tumor colonization (Fig. 4A–D), which is the rate-limiting step in metastases(24). Two weeks after injection, most of the arrested PCDH7 shRNA tumor cells either disappeared (74%) or remained as solitary cells (18%), while 70% of the extravasated control shRNA tumor cells developed into multicellular loci (≥ 4 cells). Neither PCDH7 knockdown nor transduction significantly changed in vitro tumor cell proliferation (Fig. S2E–F), prompting us to study brain microenvironment elements that may stimulate PCDH7-mediated tumor colonization.
PCDH7-mediated tumor-astrocyte interaction retains CSC self-renewal and activates PLCβ-Ca2+/CaMKII/S100A4 signaling
Immuno-reactive PCDH7 expression was detected in brain metastases in the PCDH7 shRNA group, as well as the tumor surrounding astrocytes (Fig. 3B). The re-expression of PCDH7 in the survived brain metastatic tumor cells may implicate that the tumor cells are required to adapt to express PCDH7 in order to survive in the brain. We found that when the primary MDA-MB231 cells were co-cultured with astrocytes, the expression of PCDH7 in MDA-MB231 cells was markedly up-regulated (Fig. S3A)(GSE26291). Both mouse and human astrocytes moderately express PCDH7 (Fig. S2C), and astrocytes activation was observed in brain metastases (Fig. S3B)(25). Human astrocytes facilitated retention of self-renewal for tumorspheres as evidenced by an assay of serial non-adherent passages. Tumorspheres could be propagated beyond passage 5 when co-cultured with astrocytes, but sphere cell viability was diminished by passage 2 when co-cultured with normal fibroblasts; further propagation was not possible in suspension culture (Fig. 4E). Tumorspheres and astrocytes, when grown on suspension culture, formed heterotypical spheres (Fig. 4F) that required enzymatic and mechanical disaggregation to obtain a single cell suspension before sub-culturing. This likely reflects the presence of proper cell-cell interactions among them. Application of neutralizing anti-PCDH7 antibody into the culture media not only disrupted the tumor-astrocyte heterotypical spheres, but also reduced the propagation of the spheres (Fig. 4G).
Protocadherins mediate cell-cell adhesion(26), and a recent study shows that breast and lung cancer cells express PCDH7, mediating the assembly of tumor-astrocyte gap junctions to activate astrocytes in supporting tumor growth(10). In addition, studies indicate that protocadherins are functionally important in relaying signals to cytoplasm(26). To explore whether the PCDH7 cytoplasmic signals are important to brain metastases, we transduced either a full-length PCDH7 gene (FL-PCDH7) or only the extracellular domain (EC1-3) of the PCDH7 gene (EC-PCDH7) into tumor cells (Fig. S3C), to assess tumor cell colonization and interactions with astrocytes. In animal studies, exclusive and strong astrocyte activation was seen in close proximity of cancer cells even on day 3 post cancer cell injection. We did not observe significant differences in astrocyte response in the EC-PCDH7-injected animals compared with FL-PCDH7-injected animals (Fig. S3D). However, four weeks after cell injection, 90% of the arrested or extravasated FL-PCDH7 tumor cells developed into multicellular loci (≥4 cells), while only 22% of the injected EC-PCDH7 tumor cells did (Fig. 5A–B), suggesting that the cytoplasmic signals of PCDH7 are essential for tumor colonization.
PCDH7 proteins mainly localize to cell membrane with some in the cytoplasm and nucleus of a brain metastatic tumorsphere cell (Fig. S3E). Similar to other protocadherins, PCDH7 has a small serine-rich domain in its COOH terminus that is homologous to the beta-catenin binding site of classical cadherins (Fig. S3F). Although PCDH7 binds beta-catenin directly (Fig. S3G), nuclear beta-catenin translocation and significant increases of wnt-signaling transcriptional factors (Tcf-4 and Lef-1) were not observed in the tumorsphere cells when co-cultured with astrocytes (Fig. S3H–J), suggesting that the canonical Wnt-signaling was not functional in enhancing brain metastatic tumor cell colonization.
Instead, an increased cytoplasmic Ca2+ (~ 30%) was observed independent of PCDH7 expression in the tumorsphere cells (Fig. 5C). Extensive data suggest that tumor cell proliferation is stimulated by a persistent Ca2+ increase in contrast to the transitory increase of Ca2+ that induces activation of the mitochondrial apoptotic pathway(27). PCDH7 has been shown to mediate the assembly of carcinoma–astrocyte gap junctions(10), and thus allows transmitting Ca2+ between astrocytes and brain metastatic tumor cells(25). In response to Ca2+ influx from astrocytes(25), we noted a consistently high expression and activation of phosphodiesterase beta (PLCβ) in tumorsphere cells and high expression of downstream calmodulin-dependent protein kinase II (CaMKII) and S100A4 (Fig. 5D). PLCβ catalyzes hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) to inositol 1,4,5-trisphosphate (IP3) and 1,2-diacylglycerol (DAG). Signaling pathway activation of PLCβ-IP3 or PLCβ-DAG is important for transient calcium influx-induced intracellular Ca2+ storage, which leads to activation of Ca2+/CaM-dependent kinase and nuclear transcription factors, stimulating maintenance of pluripotency and proliferation of stem cells(28). PLCβ1, CaMKII and S100A4 activations were also noted in FL-PCDH7 but not EC-PCDH7 transduced tumor cells when co-culturing with astrocytes (Fig. 5D).
We pharmacologically suppressed the PCDH7-PLCβ-Ca2+/CaMKII/S100A4 signaling by administering ET-18-OCH3 (edelfosine), a specific PLC inhibitor drug, to the MDA-MB231-Br tumorsphere xenograft mice. To recapitulate human disease where brain metastatic tumors are established before treatment, we treated mice beginning at day 11 when micro-metastatic lesions were detected in the mouse brain at day 10 (Fig. 3A). During the 15-day treatment, brain metastatic tumor growth was significantly inhibited in the ET-18-OCH3 group (P=0.04, Fig. 5E), and the formation of macro-metastases (> 50 cells) in the treated mice was inhibited by 96% at the end of the 15-day treatment (P<0.01, Fig. 5F–G), i.e., the vehicle-treated mice developed 22.5±3.5 whole-brain macro-metastases while it was only 1±0.7 in the ET-18-OCH3 treated mice. Meanwhile, activations of cellular PLCβ and tumor cell proliferation were repressed remarkably (Fig. 5H–I). Due to the limited availability of the patient derived tumorspheres, only in vitro tumorsphere treatment was performed and ET-18-OCH3 showed a dose-dependent inhibition on secondary tumorsphere formation (Fig. S4A–D). These results indicate the potential of targeting PCDH7-PLCβ-Ca2+/CaMKII/S100A4 signaling in inhibiting brain metastases.
Discussion
In this study, we identified a novel signaling pathway for brain metastatic outgrowth of TNBC and presented a new strategy for preventing or combating breast cancer metastasis via targeting organ-adaptive CSCs. We extracted a CSC population from brain metastases and demonstrated that this CSC population has an enhanced brain-adaptive capacity and engages in brain metastatic colonization, the rate-limiting event in brain metastasis(24). Further exploration of the CSC adaptation and colonization signals identified the astrocyte-involved PCDH7-PLCβ-Ca2+-CaMKII/S100A4 pathway. Suppressing the pathway’s activation by an old drug, edelfosine, showed obvious efficacy in prohibiting brain metastasis in mouse models.
Several studies suggest that CSCs of primary breast tumors are intrinsically invasive and metastatic in vivo(29,30), while others propose that migratory cells can acquire self-renewal and tumorigenic capabilities through an epithelial-mesenchymal transition (EMT)(31,32). In either case, the CSC population is characterized by adaptability to various microenvironments(33). To this end, we hypothesized that if any CSCs exist in brain metastasis, they would evolve to be more adaptive to the brain than their non-CSC counterparts. Thus, we used a “brain-friendly” culture condition (neurosphere media) to enrich the CSC population in brain metastases and demonstrated that brain metastases possess such CSC populations, i.e., they have sphere-forming capacity and are able to initiate tumor growth, reproduce original tumor heterogeneity through in vivo differentiation, and can be serially passaged in vivo. Culturing the brain-seeking cell lines in the same media also led to an enriched CSC population, and even more importantly, we were able to directly compare the functions of the paired CSCs with non-CSCs. We found that the CSC population was engaged in the brain metastatic tumor growth with much less latency and faster proliferation than the non-CSC counterparts, suggesting their intrinsic capacity in initiating brain metastasis. Through application of the CSC model, our study provides a much-needed method to study tumor adaption and explore novel metastasis mechanisms. To date, tumorspheres have been successfully cultured from various organs of cancer or metastases(34), including bone marrow, lung tissue, and pancreatic, prostate, melanoma, and ovarian cancers, suggesting an applicable generalization of using tumorsphere CSC models to study organ-specific tumor adaptation signals in metastasis.
Metastatic environment may lead to the development of organ-adaptive tumor cells, which is crucial for tumor cell thriving and surviving(35). In our study, the brain niche endows an enhanced PCDH7 expression on the survived brain metastatic CSCs, which promoted the formation of tumorspheres and retained their self-renewal. Brain cells corresponding to stromal cells are astrocytes, which are the most abundant cell type in the human brain and perform a variety of functions. It is reported that small non-coding RNAs, in particular microRNAs (miRNAs), act non-autonomously spreading from astrocytes to tumor cells by exosomes(36) or cell-to-cell contact(25,37) to enhance tumor proliferation and reduce apoptosis. Several miRNAs have recently been identified as potential regulators of PCDH7 in cancer, for example, miR-19a, miR-32, miR-124a, miR-130b, miR-148a, and miR-583(38). MiR-19a and miR-32 were also defined as glia-enriched miRNAs in brain regulating glia cell-type-specific phenotypes(39). The roles of miR-19a and miR-32 in direct or indirect up-regulation of PCDH7 in brain metastatic tumors warrant further studies. In addition, intercellular transfer of calcium, a ubiquitous second messenger, from astrocytes to tumor cells were previously examined(25), and calcium transients can increase the efficiency of gene expression and drive transcription of specific genes(40). These include PCDH family genes and genes regulating stem cell differentiation and proliferation(28).
Our research identified four aspects of PCDH7 mediated tumor-astrocyte interactions in promoting TNBC brain metastases. First, direct contacts between CSCs and astrocytes initiate the activation of PCDH7-PLCβ-Ca2+ signaling to promote tumor colonization. The cytoplasmic domain of PCDHs is structurally diverse, in contrast to the homology between classical cadherins(41), suggesting novel intracellular interactions and functions for each PCDH, but these are less studied. Our data indicate that the non-canonical Wnt/PLCβ-Ca2+ is a PCDH7 intracellular signaling event that is functionally critical to TNBC brain metastasis. Second, high-expression of PCDH7 in CSCs and astrocytes may mediate homophilic or heterophilic interactions of PCDH7 with other cadherin superfamily molecules in the same cells, neighboring cells, or both. For example, N-cadherin is highly expressed in neural tissue(41) and shares 35% sequence similarity with PCDH7 at EC1-2 domains, which could form a strand exchange trans-dimer with PCDH7 (Supplementary Table. 3). In brain metastases from lung cancer patients, Grinberg-Rashi’s group reported that N-cadherin was significantly increased(42). We postulate that PCDH7-N-cadherin interactions can propagate N-cadherin signal transduction, such as EGFR-dependent migration signaling. Our previous work confirmed that genetic or pharmacologic inhibition of EGFR had profound anti-brain metastatic effects via suppressing PLC activity(43). Third, PCDH7 expression in breast and lung cancer cells has been shown to mediate the assembly of carcinoma–astrocyte gap junctions(10) in enabling the intercellular transmission of survival and apoptotic signals by which the activated astrocytes could protect tumor cells from chemotherapy(25). Co-culturing with astrocytes induced up-regulation of many pro-survival genes (CCND1, BCL2L2 and CCNA1) and down-regulation of pro-apoptosis genes (CASP8, LATS2, FAF1 and PDCD2) in breast cancer cells (MDA-MB231) even without chemotherapeutic treatment (Fig. S3A)(GSE26291), suggesting a proactive role of astrocytes in stimulating brain tumor growth. Finally, PCDHs are proteolitically processed by γ-secretase complex, releasing soluble intracellular fragments into cytoplasm, which might have a broad range of functions locally in the cytoplasm and/or even regulate gene expression similarly to other cell-surface proteins such as Notch and N-cadherin(44,45).
Treatment for brain metastasis is an unmet medical need. Current treatment options, including radiotherapy, neurosurgery, and limited chemotherapy, frequently lead to treatment failure. Accumulated evidence has shown that elimination of CSCs is crucial in eradicating metastases(46,47). In addition, elimination of stromal support by inhibiting feedback stimulation of cancer growth has shown anti-metastatic efficacy and is the focus of many emerging therapies(48). As such, in our tumorsphere CSC animal studies, edelfosine almost completely eradicated the macro-metastases in the mouse brain through inhibition of cell proliferation and breaching the astrocyte-tumor interplay. Edelfosine is a synthetic alkyl-lysophospholipid and a selective PLC inhibitor. It has been trialed in a phase II study for the treatment of brain cancer and showed encouraging results in stopping the tumor growth and a considerable improvement in the quality of life of the patients (US Patent 6514519). The drug incorporates into the cell membrane and does not target the DNA, thus it causes selective apoptosis in tumor cells, sparing healthy cells(49). While the existing treatment strategies for brain metastases are toxic to healthy tissues and significantly affect the patients’ quality of life, our results indicate that further study of edelfosine as a potential treatment option for TNBC brain metastasis is warranted.
Supplementary Material
Acknowledgments
We thank all members of the Wong laboratory at Houston Methodist for discussions and assistance with data analysis, Drs. Neal Copeland and Jeffrey Rosen provided helpful comments for the manuscript, Drs. Rebecca Danforth and James Mancuso for their proofreading skills, and Drs. Patricia Steeg and Joan Massague generously provided MDA-MB231-Br and CN34-Br cell lines. All microscopic imaging experiments were performed at Houston Methodist Research Institute’s Advanced Cellular and Tissue Microscope Core Facility. We acknowledge the computational time funding support from the Texas Advanced Computing Center (TACC; Project ID: TG-MCB110130) at the University of Texas in Austin and BlueBioU (IBM POWER 7 Bioscience Computing Core at Rice University) to access super-computing resources. This research was funded by NIH U54 CA149196, NIH R01 CA121225, and John S. Dunn Research Foundation grants to S.T.Wong, and R01-CA183878 to X.HF.Zhang.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Qiu J, Xue X, Hu C, Xu H, Kou D, Li R, et al. Comparison of Clinicopathological Features and Prognosis in Triple-Negative and Non-Triple Negative Breast Cancer. J Cancer. 2016;7(2):167–73. doi: 10.7150/jca.10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Owonikoko TK, Arbiser J, Zelnak A, Shu HK, Shim H, Robin AM, et al. Current approaches to the treatment of metastatic brain tumours. Nat Rev Clin Oncol. 2014;11(4):203–22. doi: 10.1038/nrclinonc.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leone JP, Leone BA. Breast cancer brain metastases: the last frontier. Exp Hematol Oncol. 2015;4:33. doi: 10.1186/s40164-015-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niikura N, Hayashi N, Masuda N, Takashima S, Nakamura R, Watanabe K, et al. Treatment outcomes and prognostic factors for patients with brain metastases from breast cancer of each subtype: a multicenter retrospective analysis. Breast Cancer Res Treat. 2014;147(1):103–12. doi: 10.1007/s10549-014-3090-8. [DOI] [PubMed] [Google Scholar]
- 5.Oskarsson T, Batlle E, Massague J. Metastatic stem cells: sources, niches, and vital pathways. Cell Stem Cell. 2014;14(3):306–21. doi: 10.1016/j.stem.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luzzi KJ, MacDonald IC, Schmidt EE, Kerkvliet N, Morris VL, Chambers AF, et al. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am J Pathol. 1998;153(3):865–73. doi: 10.1016/S0002-9440(10)65628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kienast Y, von Baumgarten L, Fuhrmann M, Klinkert WE, Goldbrunner R, Herms J, et al. Real-time imaging reveals the single steps of brain metastasis formation. Nat Med. 2010;16(1):116–22. doi: 10.1038/nm.2072. [DOI] [PubMed] [Google Scholar]
- 8.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525(7568):256–60. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015 doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Q, Boire A, Jin X, Valiente M, Er EE, Lopez-Soto A, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. 2016;533(7604):493–8. doi: 10.1038/nature18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tropepe V, Sibilia M, Ciruna BG, Rossant J, Wagner EF, van der Kooy D. Distinct neural stem cells proliferate in response to EGF and FGF in the developing mouse telencephalon. Dev Biol. 1999;208(1):166–88. doi: 10.1006/dbio.1998.9192. [DOI] [PubMed] [Google Scholar]
- 12.Zhao H, Cui K, Nie F, Wang L, Brandl MB, Jin G, et al. The effect of mTOR inhibition alone or combined with MEK inhibitors on brain metastasis: an in vivo analysis in triple-negative breast cancer models. Breast Cancer Res Treat. 2012;131(2):425–36. doi: 10.1007/s10549-011-1420-7. [DOI] [PubMed] [Google Scholar]
- 13.Rota LM, Lazzarino DA, Ziegler AN, LeRoith D, Wood TL. Determining mammosphere-forming potential: application of the limiting dilution analysis. J Mammary Gland Biol Neoplasia. 2012;17(2):119–23. doi: 10.1007/s10911-012-9258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–9. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu W, He Q, Li X, Zhang X, Lu A, Ge R, et al. Long-term cultured human neural stem cells undergo spontaneous transformation to tumor-initiating cells. Int J Biol Sci. 2011;7(6):892–901. doi: 10.7150/ijbs.7.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sihto H, Lundin J, Lundin M, Lehtimaki T, Ristimaki A, Holli K, et al. Breast cancer biological subtypes and protein expression predict for the preferential distant metastasis sites: a nationwide cohort study. Breast Cancer Res. 2011;13(5):R87. doi: 10.1186/bcr2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jhanwar-Uniyal M, Labagnara M, Friedman M, Kwasnicki A, Murali R. Glioblastoma: molecular pathways, stem cells and therapeutic targets. Cancers (Basel) 2015;7(2):538–55. doi: 10.3390/cancers7020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo L, Fan D, Zhang F, Price JE, Lee JS, Marchetti D, et al. Selection of brain metastasis-initiating breast cancer cells determined by growth on hard agar. Am J Pathol. 2011;178(5):2357–66. doi: 10.1016/j.ajpath.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66(19):9339–44. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 20.Wilhelm I, Molnar J, Fazakas C, Hasko J, Krizbai IA. Role of the blood-brain barrier in the formation of brain metastases. Int J Mol Sci. 2013;14(1):1383–411. doi: 10.3390/ijms14011383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabrera MC, Hollingsworth RE, Hurt EM. Cancer stem cell plasticity and tumor hierarchy. World J Stem Cells. 2015;7(1):27–36. doi: 10.4252/wjsc.v7.i1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato N, Sanjuan IM, Heke M, Uchida M, Naef F, Brivanlou AH. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol. 2003;260(2):404–13. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki ST. Recent progress in protocadherin research. Exp Cell Res. 2000;261(1):13–8. doi: 10.1006/excr.2000.5039. [DOI] [PubMed] [Google Scholar]
- 24.Termini J, Neman J, Jandial R. Role of the neural niche in brain metastatic cancer. Cancer Res. 2014;74(15):4011–5. doi: 10.1158/0008-5472.CAN-14-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Q, Balasubramanian K, Fan D, Kim SJ, Guo L, Wang H, et al. Reactive astrocytes protect melanoma cells from chemotherapy by sequestering intracellular calcium through gap junction communication channels. Neoplasia. 2010;12(9):748–54. doi: 10.1593/neo.10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank M, Kemler R. Protocadherins. Curr Opin Cell Biol. 2002;14(5):557–62. doi: 10.1016/s0955-0674(02)00365-4. [DOI] [PubMed] [Google Scholar]
- 27.Jaffe LF. A calcium-based theory of carcinogenesis. Adv Cancer Res. 2005;94:231–63. doi: 10.1016/S0065-230X(05)94006-2. [DOI] [PubMed] [Google Scholar]
- 28.Tonelli FM, Santos AK, Gomes DA, da Silva SL, Gomes KN, Ladeira LO, et al. Stem cells and calcium signaling. Adv Exp Med Biol. 2012;740:891–916. doi: 10.1007/978-94-007-2888-2_40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, et al. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med. 2009;13(8B):2236–52. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci U S A. 2010;107(42):18115–20. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang X, Cai Y, Liu J, Wang Z, Wu Q, Zhang Z, et al. Twist2 contributes to breast cancer progression by promoting an epithelial-mesenchymal transition and cancer stem-like cell self-renewal. Oncogene. 2011;30(47):4707–20. doi: 10.1038/onc.2011.181. [DOI] [PubMed] [Google Scholar]
- 32.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Csermely P, Hodsagi J, Korcsmaros T, Modos D, Perez-Lopez AR, Szalay K, et al. Cancer stem cells display extremely large evolvability: alternating plastic and rigid networks as a potential Mechanism: network models, novel therapeutic target strategies, and the contributions of hypoxia, inflammation and cellular senescence. Semin Cancer Biol. 2015;30:42–51. doi: 10.1016/j.semcancer.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Tirino V, Desiderio V, Paino F, De Rosa A, Papaccio F, La Noce M, et al. Cancer stem cells in solid tumors: an overview and new approaches for their isolation and characterization. FASEB J. 2013;27(1):13–24. doi: 10.1096/fj.12-218222. [DOI] [PubMed] [Google Scholar]
- 35.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9(4):239–52. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527(7576):100–04. doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menachem A, Makovski V, Bodner O, Pasmanik-Chor M, Stein R, Shomron N, et al. Intercellular transfer of small RNAs from astrocytes to lung tumor cells induces resistance to chemotherapy. Oncotarget. 2016;7(11):12489–504. doi: 10.18632/oncotarget.7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aakula A, Kohonen P, Leivonen SK, Makela R, Hintsanen P, Mpindi JP, et al. Systematic Identification of MicroRNAs That Impact on Proliferation of Prostate Cancer Cells and Display Changed Expression in Tumor Tissue. Eur Urol. 2016;69(6):1120–8. doi: 10.1016/j.eururo.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 39.Jovicic A, Roshan R, Moisoi N, Pradervand S, Moser R, Pillai B, et al. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J Neurosci. 2013;33(12):5127–37. doi: 10.1523/JNEUROSCI.0600-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392(6679):933–6. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 41.Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20(23):3199–214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 42.Grinberg-Rashi H, Ofek E, Perelman M, Skarda J, Yaron P, Hajduch M, et al. The expression of three genes in primary non-small cell lung cancer is associated with metastatic spread to the brain. Clinical Cancer Research. 2009;15(5):1755–61. doi: 10.1158/1078-0432.CCR-08-2124. [DOI] [PubMed] [Google Scholar]
- 43.Nie F, Yang J, Wen S, An YL, Ding J, Ju SH, et al. Involvement of epidermal growth factor receptor overexpression in the promotion of breast cancer brain metastasis. Cancer. 2012;118(21):5198–209. doi: 10.1002/cncr.27553. [DOI] [PubMed] [Google Scholar]
- 44.Bonn S, Seeburg PH, Schwarz MK. Combinatorial expression of alpha- and gamma-protocadherins alters their presenilin-dependent processing. Mol Cell Biol. 2007;27(11):4121–32. doi: 10.1128/MCB.01708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buchanan SM, Schalm SS, Maniatis T. Proteolytic processing of protocadherin proteins requires endocytosis. Proc Natl Acad Sci U S A. 2010;107(41):17774–9. doi: 10.1073/pnas.1013105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen K, Huang YH, Chen JL. Understanding and targeting cancer stem cells: therapeutic implications and challenges. Acta Pharmacol Sin. 2013;34(6):732–40. doi: 10.1038/aps.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dragu DL, Necula LG, Bleotu C, Diaconu CC, Chivu-Economescu M. Therapies targeting cancer stem cells: Current trends and future challenges. World J Stem Cells. 2015;7(9):1185–201. doi: 10.4252/wjsc.v7.i9.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engels B, Rowley DA, Schreiber H. Targeting stroma to treat cancers. Semin Cancer Biol. 2012;22(1):41–9. doi: 10.1016/j.semcancer.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mollinedo F, Gajate C, Martin-Santamaria S, Gago F. ET-18-OCH3 (edelfosine): a selective antitumour lipid targeting apoptosis through intracellular activation of Fas/CD95 death receptor. Curr Med Chem. 2004;11(24):3163–84. doi: 10.2174/0929867043363703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


