Figure 1. Alignment and phylogenetic tree of c8orf46 orthologs.
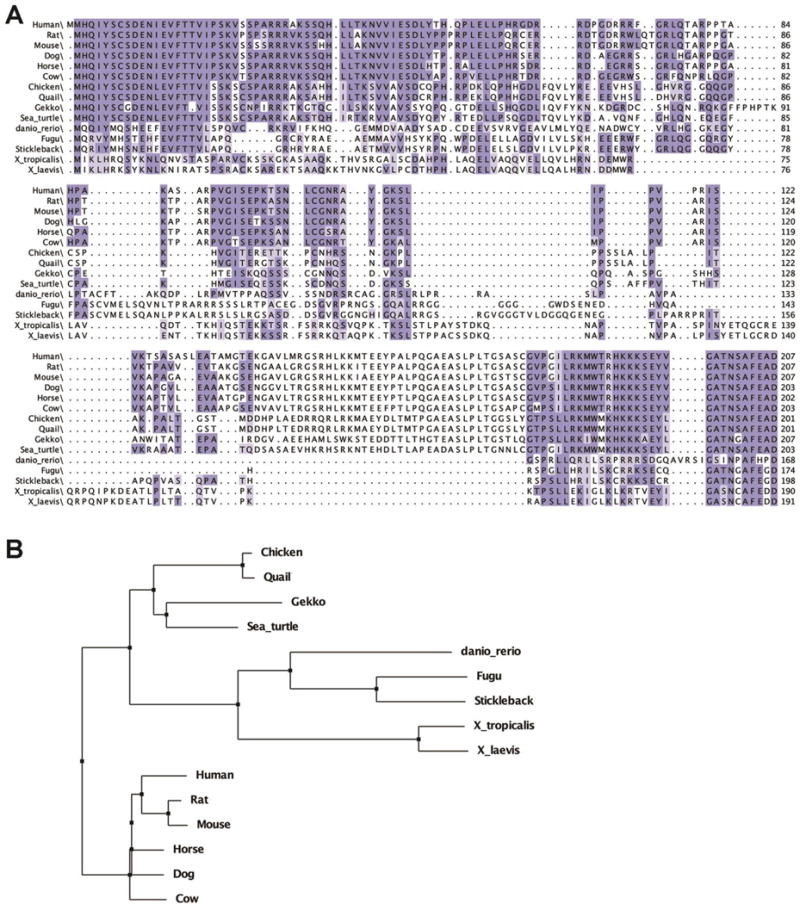
(A) The predicted protein sequence encoded by the Xenopus laevis gene c8orf46.L (vxn) was aligned to predicted protein sequences encoded by c8orf46 orthologs from other vertebrate species (see methods for accession numbers) using the webPRANK program in Jalview2 (Waterhouse et al., 2009). Residues conserved in at least 70% of the aligned sequences are indicated with dark blue shading while light blue shading indicates residues that are 50-69% conserved. (B) The webPRANK alignment was used to generate a phylogenetic tree using BLOSUM62.
