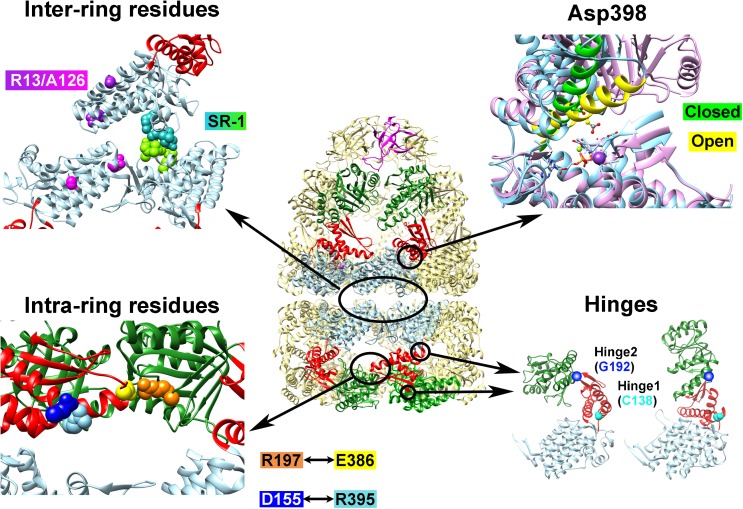
Fig. 2.
Examples of GroEL mutants where the functional mechanism has been “reprogrammed”. Depicted at center is the cis–GroEL–GroES complex from 1SVT; arranged around this depiction are four panels which show the locations of various amino acid residues that were mutated to induce functional modifications to GroEL. The Inter-ring residues panel shows the four amino acid residues that are altered to form the single-ring GroEL mutant SR-1 in green and turquoise, and Arg13 and Ala126, which were altered to negate the negative cooperativity between GroEL rings, are shown in magenta and purple in their respective subunits. In the Asp398 panel, Helix M, which orients this residue toward the ATP binding site upon nucleotide binding, is depicted in green for the closed form and yellow in the open form. Asp398 is depicted in ball and stick form. This panel was generated by fitting the equatorial domains of the open and closed forms of the GroEL subunits in PDBID 1SVT using the MatchMaker tool in UCSF Chimera (Pettersen et al. 2004). The Intra-ring residues panel shows the location of Arg197 (orange), Glu386 (yellow), Asp155 (blue), and Arg395 (light blue). The Hinges panel shows the two residues that were modified to confer temperature sensitivity (Cys138, cyan) and substrate binding affinity (Gly192, dark blue)