Abstract
The need for accurate description of protein behavior in solution has gained importance in various fields, including biophysics, biochemistry, structural biology, drug discovery, and antibody drugs. To achieve the desired accuracy, multiple precise analyses should be performed on the target molecule, compared, and effectively combined. This review focuses on the combination of multiple analyses in solution: size-exclusion chromatography (SEC), multi-angle light scattering (MALS), small-angle X-ray scattering (SAXS), analytical ultracentrifugation (AUC), and their complementary methods, such as atomic force microscopy (AFM) and mass spectrometry (MS). We also discuss the comparison between the determined molar mass value of not only the standard proteins, but of a target molecule tubulin and its depolymerizing protein, KIF2, as an example. The comparison of the estimated molar mass value from the different methods provides additional information about the target molecule, because the value reflects the dynamically changing states of the target molecule in solution. The combination and integration of multiple methods will permit a deeper understanding of protein dynamics in solution.
Keywords: Size-exclusion chromatography (SEC), Multi-angle light scattering (MALS), Small-angle X-ray scattering (SAXS), Analytical ultracentrifugation (AUC), Microtubule, Kinesin
Introduction
Proteins form dynamic structures and work efficiently to perform their functions. The proper conformation of the protein complexes in solution is the basis of their physiological function. In solution, proteins fulfill their function through the use of their multi-dimensional features, such as conformational change, interaction, complex formation, post-translational modification, degradation, and the change of their molar mass value. Therefore, the analysis of the significant change in molar mass value (molecular size) and their conformation delivers critical insights into the physiological mechanism of the function of the target molecule. When considering protein function through the proper conformation of the protein complex in solution, their precise analyses should be conducted in solution.
Over the last few decades, many improvements have occurred in the fields of protein research; theory, methods, measurement equipment, and computers for calculation. This review focuses on the effective approaches presented by these recent advances, which combine multiple methods for protein analysis in solution: size-exclusion chromatography (SEC), multi-angle light scattering (MALS), small-angle X-ray scattering (SAXS), and analytical ultracentrifugation (AUC). In addition, this review also introduces their complementary methods, such as mass spectrometry (MS) and high-speed atomic force microscopy (HS-AFM). In particular, separation by SEC was greatly improved and coupled efficiently with other methods in line. The combination and integration of multiple methods can provide a deeper understanding of protein dynamics in solution.
High-resolution size-exclusion chromatography (HiRes SEC)
SEC (gel-filtration chromatography) is an established method for the separation and analysis of molecules based on their hydrodynamic volume. Conventional SEC provides good separation of large molecules from small molecules, and also provides valid information about the molecular mass distribution and molecular weight (Mw). However, even for molecules of the same size, some proteins may display a low molecular weight with a loosely packed structure, whereas other proteins may show a high molecular weight with a densely packed structure. Thus, the molecular weight cannot be determined by only the elution position. SEC is limited by the working range for each column, in which the upper limit is defined as the exclusion limit by the void volume and the lower limit is defined as the permeation limit determined by the column volume. Because of these limitations, conventional SEC columns did not provide efficient separation in the middle range of molecular weight approximately 100–200 kDa, for example. Another limitation is the column pressure limit, which restricts the performance at higher flow rates. Because using a longer SEC column results in a much longer time for separation at lower flow rates, unstable samples cannot be analyzed within their retention time. An SEC separation technique that can combine a higher resolution and a shorter runtime is eagerly awaited.
Recently, the maximum pressure limit of the Superdex 200 Increase 10/300 column (GE Healthcare) was improved to 3.0 MPa, compared with that of the conventional Superdex 200 Column (1.5 MPa). When twin Superdex 200 Increase 10/300 columns were connected in tandem (Fig. 1a), fresh samples can be separated at higher resolution under 3.0 MPa pressure, which corresponds to a flow rate of approximately 0.4 mL/min for general buffers. These improvements in SEC columns are able to overcome some of the bottlenecks of protein analyses. The first advantage is the increased resolution for sample separation: for example, high-resolution SEC (HiRes SEC) can separate ovalbumin solution into its monomer, dimer, and tetramer populations (Fig. 1b). This improvement allows SEC separation to obtain mono-disperse eluate and make it suitable for subsequent inline analyses. The second advantage is the shorter runtime for separation: most high-resolution separations can be completed within 100 min, which enables the freshly eluted samples to be analyzed in combination with other equipment.
Fig. 1.
High-resolution size-exclusion chromatography (HiRes SEC) displays good separation. a HiRes SEC composed of a tandem connection of twin Superdex 200 Increase 10/300 columns (GE Healthcare). b HiRes SEC displays a good separation of ovalbumin in the monomeric, dimeric, and tetrameric forms. The column was equilibrated with assay buffer (20 mM PIPES, 200 mM KCl, 1 mM MgCl2) and assayed at a flow rate of 0.4 mL/min on an ÄKTA system (GE Healthcare)
Combination of HiRes SEC with other methods
As described above, HiRes SEC can provide sufficiently a high resolution for the separation of ovalbumin into the monomeric, dimeric, and tetrameric mono-disperse fractions within 100 min (Fig. 1b). This separation is effective for further analyses, which require the sample to be mono-disperse in solution in order to ensure the accuracy of the analysis. In fact, scattering-based approaches, such as MALS and SAXS, are typical cases for improvement by combination with a HiRes SEC system.
MALS is one of the most reliable techniques for the determination of the absolute molar mass and rms radius r g in solution (Wyatt 1998). When a protein standard mixture consisting of ferritin (440 kDa), aldolase (158 kDa), ovalbumin (44 kDa), and RNase (13.7 kDa) is separated into mono-disperse fractions by HiRes SEC (Fig. 2a), each eluate can be directly analyzed in-line by the subsequent application of MALS (Fig. 2b) and refractive index (RI) detection (Fig. 2c) (Wyatt 1998). HiRes SEC-RI-MALS analysis indicated the protein molecular weights as 671 kDa for ferritin, 161 kDa for aldolase, 50 kDa for ovalbumin, and 19.8 kDa for RNase. Because light scattering is sensitive to protein aggregation, the HiRes SEC-MALS system works effectively to separate and detect the aggregates. Because aggregates and fragments exist in antibody drugs, they may potentially affect immunogenicity and potency. Thus, the SEC-MALS approach is also a good option for the analysis and validation of antibody drugs (Lu et al. 2013).
Fig. 2.
Combination of HiRes SEC and the refractive index (RI)-multi-angle light scattering (MALS) system. a Protein standard mixture (500 μL) containing ferritin (estimated Mw: 440 kDa), aldolase (158 kDa), ovalbumin (44 kDa), and RNase (13.7 kDa) (100 μg per protein) was separated as mono-disperse fractions by HiRes SEC. Each eluate was directly analyzed by the refractive index (RI) and Dawn Heleos II 18-angle MALS detectors (RI-MALS) (Wyatt). b HiRes SEC-MALS indicated the molecular weight of each protein as 671 kDa for ferritin, 161 kDa for aldolase, 50 KDa for ovalbumin, and 19.8 kDa for RNase. Aggregates at void volume show a large peak. c HiRes SEC-RI detector. The column was equilibrated with assay buffer (20 mM PIPES, 200 mM KCl, 1 mM MgCl2) and assayed at a flow rate of 0.4 mL/min
SAXS analysis is another scattering-based approach for the analysis of proteins in solution. The SAXS technique provides information about protein structure and dynamics, such as r g, Dmax, and the outline of the molecule in solution. Similar to MALS, SAXS measurements also require the sample to be mono-disperse in solution; additionally, as protein solutions are usually highly sensitive to the X-ray damage, protein aggregation may occur and reduce the accuracy of SAXS measurements. In conventional static SAXS measurements, the sample is held in the sample cell holder for a long exposure time; thus, only proteins resistant to X-ray damage can be analyzed by this technique. Recently, HiRes SEC-SAXS has overcome this problem. In HiRes SEC-SAXS analysis, a HiRes SEC column with an HPLC system is connected with the following flow-cell for the SAXS measurement and the eluate is directly analyzed by SAXS at the appropriate beamline in the synchrotron (Igarashi et al. 2013). In HiRes SEC-SAXS measurement, the injected sample is separated on a HiRes SEC. The mono-disperse eluate directly flows into a flow-cell holder and serial scattering images are taken over a certain exposure time. Because the fresh sample continuously flows into the cell holder, the X-ray damage to the protein is greatly reduced. The scattering images are circularly averaged and converted to one-dimensional scattering intensity data and data processing is performed. Although the injected sample is diluted during SEC separation, high S/N scattering data enough to outline the overall structure can be obtained by X-ray at the synchrotron. Additional improvements to collect the data with a much higher S/N intensity from a low concentration of samples would represent a future advancement of this technique.
Validation and further analyses by theoretically independent approaches
As described above, HiRes SEC-MALS and HiRes SEC-SAXS are dependent on SEC separation. To validate the molecular size in solution, multiple theoretically independent approaches are effective. A cross-check by using multiple methods should confirm the results of HiRes SEC-based analyses in solution. In addition, comparisons between different methods sometimes provide additional information about the target molecule in solution, because the value that is measured in each method can reflect the situation of the target molecule, such as conformation, folding, activity, equilibrium, and dissociation.
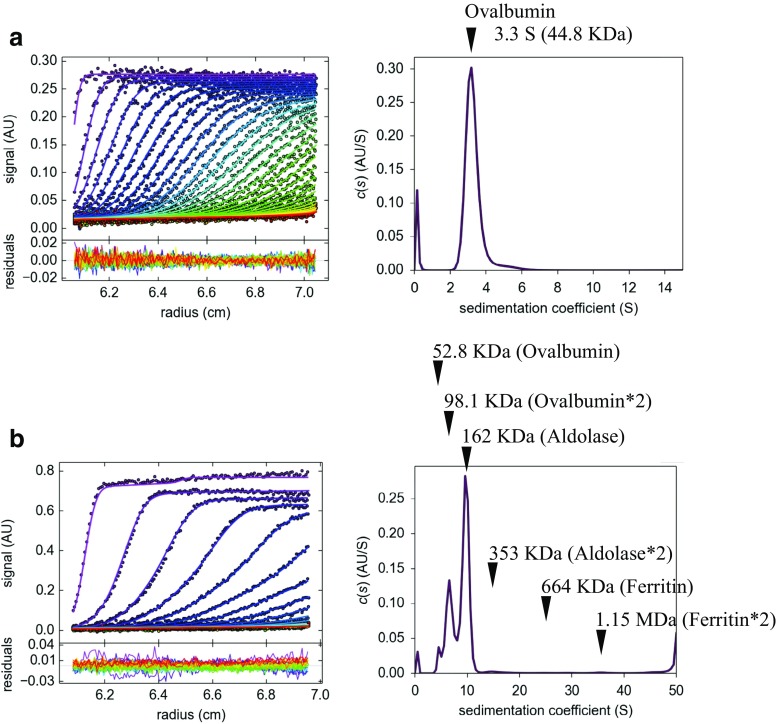
AUC would be the first choice for the validation of the molecular size in solution. Compared with the SEC-MALS system, AUC can simultaneously conduct the separation and analysis in solution. AUC can analyze target molecules with a wide range of molecular weights, from peptides to macromolecules (Schuck 2013; Arisaka and Kanamaru 2013; Uchiyama et al. 2016; Arisaka et al. 2016). Sedimentation velocity experiments are usually conducted at 20 °C by using a specified analytical ultracentrifuge. Concentration gradients are usually measured by UV absorption at 280 nm. The partial specific volumes of the proteins, buffer density, and viscosity are computed by the SEDNTERP program (Laue et al. 1992). The distribution functions of the sedimentation coefficients, c(s), are computed by the SEDFIT program (Schuck 2000), on the assumption that the frictional ratio is common to all molecular species. The values of c(s) are converted to c(M), the distribution of the molecular weights, on the basis of the Svedberg equation, which is implemented in SEDFIT. The molecular size of ovalbumin was determined by AUC as 44.8 kDa (Fig. 3a), which corresponds to the estimated value, 44 kDa. The standard mixture solution was also analyzed by AUC. The determination of the molecular size of each standard protein in the mixture solution is shown in Fig. 3b. Interestingly, at high concentrations, the determined value might be affected at equilibrium through specific dimerization and dissociation. In fact, the dimerized components were also observed in AUC measurements (Fig. 3b).
Fig. 3.
Analytical ultracentrifugation (AUC) determines the molecular mass in solution. a Ovalbumin was analyzed by AUC. b A standard mixture containing ferritin (estimated Mw: 440 kDa), aldolase (158 kDa), ovalbumin (44 kDa), and RNase (13.7 kDa) (100 μg for each protein) was analyzed by AUC
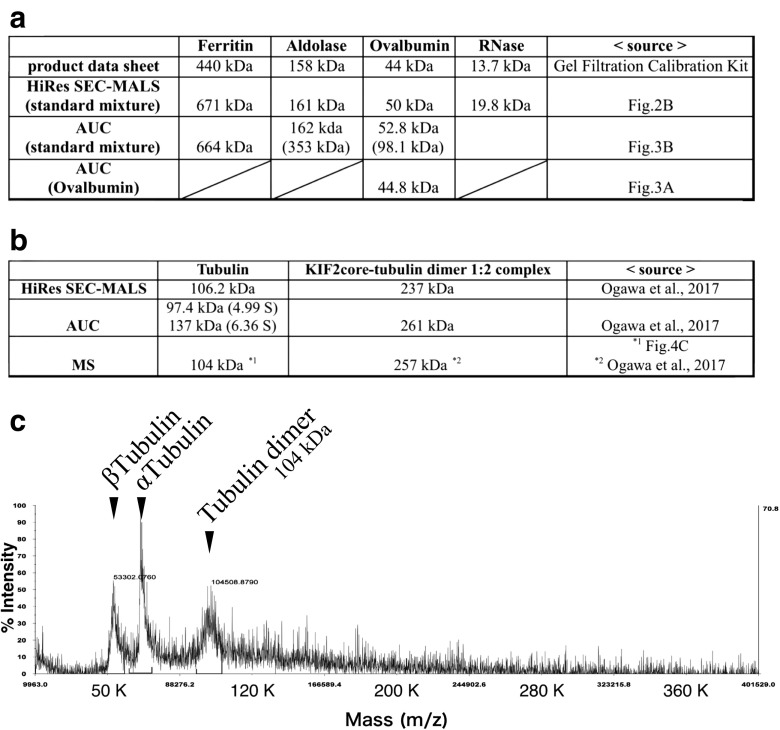
The analyses of molecular size in solution sometimes provide ambiguous results because these values reflect the dynamically changing states in solution. In these cases, the absolute molecular value of the target molecule can be determined by using a combination of chemical crosslinking technology and mass spectrometry, which is called crosslink mass spectrometry (X-link MS). To measure large proteins, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is useful because it mainly produces monovalent ions of protein/peptides. When the protein complex is analyzed by MALDI-TOF, strong lasers induce the dissociation of the protein complex. Therefore, the complex should be fixed by a chemical crosslinker. In X-link MALDI-TOF MS measurements, not only the purity and the mono-dispersity of the protein sample, but also the selection of the crosslinker and the matrix are critical for effective measurements. The MALDI-TOF spectrum of the tubulin heterodimer complex that was crosslinked by a zero-length crosslinker, EDC, before measurement is shown in Fig. 4c. In MALDI-TOF, the 104-kDa MS spectra of tubulin heterodimer was monitored in addition to the dissociated (α-/β-)tubulin monomers (Fig. 4c). MALDI-TOF measurement is conducted not in solution, but in vacuo; however, the comparison of its absolute value with the other values in solution delivers critical insights into the function of the target molecule. In addition, protein–protein interactions can also be analyzed by X-link MS. In this case, the complex is crosslinked, digested into peptides, and analyzed by LC-MS2/MS3. Sample purity and crosslinking conditions are naturally critical factors, which should be optimized to avoid over-crosslinking and the production of artifacts.
Fig. 4.
Comparison of molar mass values estimated by multiple analyses. a A comparison of the estimated and measured values of the standard proteins: ferritin, aldolase, ovalbumin, and RNase. The molecular weight of the standard proteins was estimated independently by HiRes SEC-MALS and AUC. b Summary of the measured value of tubulin and the KIF2-tubulin 1:2 complex. The molecular weight of tubulin, KIF2-tubulin 1:2 complex was measured by HiRes SEC-MALS, AUC, and crosslink (X-link) matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). c The molecular weight of tubulin was analyzed by MALDI-TOF MS (ABI 4700 MALDI-TOF mass spectrometer, SCIEX) in linear mode with a 2,5-dihydroxybenzoic acid (DHB) matrix (WAKO 046-02262)
For example, we recently reported that one core domain of the microtubule depolymerizing machine KIF2 formed a 1:2 complex with tubulin dimers (1:2 complex) (Ogawa et al. 2017). To determine the molecular mass of the KIF2–tubulin complex in solution, the reaction mixture of the KIF2–tubulin complex was applied onto the HiRes SEC, and separation was performed in-line with MALS by using an HPLC system equipped with Dawn Heleos II 18-angle MALS detectors (Wyatt). The estimated value of the 1:2 complex by HiRes SEC-MALS was 237 kDa in solution. In contrast, AUC estimated the molecular mass as 261 kDa in solution (Ogawa et al. 2017) (Fig. 4a). Therefore, in order to determine the exact molecular mass, a 1:2 complex fraction of HiRes SEC was crosslinked with the zero-length crosslinker EDC. The crosslinked sample was analyzed by MALDI-TOF in linear mode. This measurement directly suggested that the 1:2 complex was exactly 257 kDa and is composed of one KIF2 (51 kDa) and two tubulin dimers (103 kDa) (1:2 complex). Interestingly, the 1:2 complex size of 257 kDa perfectly reflected the sum of its components: 51 kDa (KIF2 core construct including the T7-tag and His-tag) and two sets of 103 kDa (tubulin dimer) (1:2 complex) (Ogawa et al. 2017). To further analyze the interaction, the 1:2 complex was crosslinked with EDC or DSSO, trypsinized, and analyzed by LC-MS. X-link MS indicated a specific interaction between KIF2 and the tubulin dimers (Ogawa et al. 2017).
Comparison and integration of multiple analyses
In solution, the protein does not always appear to have the same molecular mass; this has been a long-standing and important topic for discussion. The molecular mass values reflect not only the complex’s molecular weight, but also its conformation and the equilibrium between the molecules near the moving boundary in solution (Gilbert and Gilbert 1973; Ishii et al. 2012). In the comparison of the standard proteins, aldolase and ovalbumin show both a monomer and dimer when analyzed by AUC (Fig. 4a). This indicated that these proteins may form a dimer in the moving boundary of a highly concentrated mixture during AUC analysis. Ferritin is an iron storage protein whose molar mass is approximately 480 kDa without irons (apoferritin). Interestingly, ferritin shows different molar mass in AUC and MALS. The increase in molar mass to 600–900 KDa is reported to be due to its iron contents (Ghirlando et al. 2016). The tubulin dimer is a typical protein unit that exists in equilibrium between the polymerized microtubule and the depolymerized tubulin dimer in solution. Interestingly, the tubulin dimer (103 kDa) also displays two different molecular species in AUC analysis with sedimentation coefficients of 4.99 S (97.4 kDa) and 6.36 S (137 kDa) (Ogawa et al. 2017). In contrast, HiRes SEC-MALS and X-link MS displayed one species of 104–106 kDa in molecular weight (Fig. 4b, c). Because the AUC is sensitive to the conformation and the equilibrium of the samples, we believe it is possible that the tubulin heterodimer can form different conformations in solution or is involved in the equilibrium of the dynamic instability between polymerization and depolymerization in moving boundary conditions during AUC. Given that the tubulin heterodimer is composed of α- and β-tubulin, we think that the two different values estimated by AUC (97.4 and 137 kDa) indicated two different conformations of the tubulin heterodimer.
Additional approaches
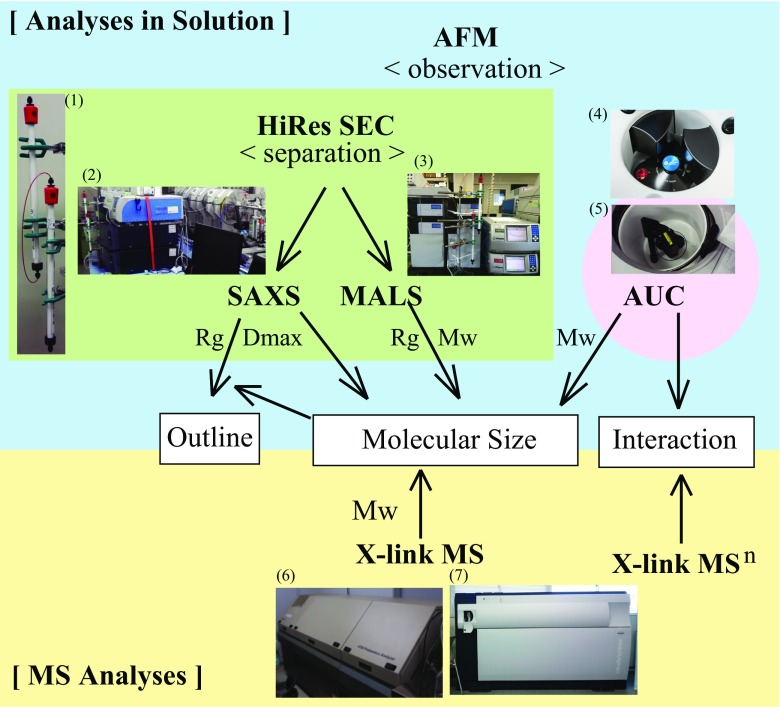
High-speed atomic force microscopy (HS-AFM) is a straightforward approach for the observation of the dynamic proteins at the single molecule level in solution; the technique can observe the movement of the protein in solution at 10–16 frames/s (Ando 2017). This can provide real-time imaging of the dynamic changes in the target protein, such as protein–protein interaction, flexibility, and the conformational change (Kodera et al. 2010; Uchihashi et al. 2011). In order to make the best use of HS-AFM, the target sample should be precisely analyzed before observation; molecular size, mono-dispersity, function, and stability in solution. In our recent study, we observed that the large complex of KIF2–tubulin was dissociated from the tubulin protofilament by using HS-AFM. This observation confirmed our subsequent analyses on the molecular mass of the KIF2–tubulin complex (Ogawa et al. 2017) (Fig. 5).
Fig. 5.
Schematic diagram of the combination of multiple analyses of the target protein. Observation: high-speed atomic force microscopy (HS-AFM) directly monitors the protein sample in solution. Separation: HiRes SEC separates the protein sample and elutes as a mono-disperse fraction. Molecular size determination in solution: based on the separation of HiRes SEC, MALS and SAXS analyses are applied to the mono-disperse sample, to provide accurate measurement of the molecular size in solution. Molecular size determination in vacuo: the peak fraction eluted from HiRes SEC can be analyzed by MALDI-TOF MS to obtain the absolute value. Outline: SAXS analysis provides the outline of the target molecule based on SAXS data and the exact molar mass value. Interaction: the interaction within the complex can be analyzed by X-link MSn. Photos: (1) HiRes SEC composed of a tandem connection of twin Superdex 200 Increase 10/300 columns (GE Healthcare); (2) HiRes SEC-SAXS at beamline BL-15A2 of the Photon Factory of the High Energy Accelerator Research Organization (KEK, Tsukuba, Japan); (3) HPLC (Shimadzu) system equipped with Dawn Heleos II 18-angle MALS detectors (Wyatt); (4) Optima AUC (Beckman Coulter); (5) ProteomeLab XL-A AUC (Beckman Coulter); (6) ABI 4700 MALDI-TOF MS (SCIEX); (7) ultrafleXtreme MALDI-TOF MS (Bruker Daltonics)
Application of the combination of multiple analyses in solution
In a recent study (Ogawa et al. 2017), we focused on the transitional large complex of KIF2–tubulin in solution, as described above. KIF2 is one of the protein sub-family members in the kinesin superfamily motor proteins (KIFs), which contains a highly conserved motor domain composed of ATP-binding and microtubule (MT) binding sites (Hirokawa 1998). Although most KIFs bind and move along MTs, the KIF2 protein has a unique ability to depolymerize MTs during ATP hydrolysis (Walczak et al. 1996; Desai et al. 1999). This unique activity was partially explained by the specific structural features revealed by the X-ray crystallographic structure of KIF2 in previous reports (Ogawa et al. 2004; Shipley et al. 2004). However, KIF2 accomplishes its function on the dynamic tubulin protofilaments in solution. Therefore, the conformation of KIF2 with the tubulin dimer during the MT depolymerization should be clarified in solution. To elucidate the transitional KIF2–tubulin conformation in solution, multiple approaches have been applied, including HS-AFM, HiRes SEC, HiRes SEC-MALS, HiRes SEC-SAXS, AUC, and MS.
HS-AFM directly monitored the depolymerization in solution and suggested that the large KIF2–tubulin complex is catalytically dissociated in an ATP-dependent manner (Fig. 5, <observation>). HiRes SEC elucidated the critical state during ATP-hydrolysis for depolymerization, which indicated that KIF2 and the tubulin dimer form the large transitional complex in the middle of the pre-hydrolysis state. In addition, HiRes SEC displayed a shifted peak of the large complex successfully separated from the next peak of the smaller complex. This separation predominantly contributed to the accurate measurement of the large complex in solution (Fig. 5, <separation>). Based on the results of HiRes SEC analysis and separation, HiRes SEC-MALS was applied to the KIF2–tubulin complex. The large KIF2–tubulin complex was estimated to have a molecular weight of 237 kDa by MALS (Fig. 5, “Molecular Size”). The same sample was analyzed by AUC, which suggested that the large complex was 261 kDa (Fig. 5, “Molecular Size”). To obtain the absolute value, the peak fraction of the large complex from HiRes SEC was analyzed by X-link MALDI-TOF MS. The molecular weight was exactly 257 kDa (Fig. 5, “Molecular Size”), which corresponded perfectly to the sum of its components: 51 kDa (KIF2) and two sets of 103 kDa (tubulin dimer) (1:2 complex). This measurement directly suggested that one KIF2 forms a large complex with two sets of tubulin dimers in the middle of the pre-hydrolysis state. To reveal the structural arrangement of the 1:2 complex in solution, HiRes SEC-SAXS was conducted at the synchrotron (Igarashi et al. 2013; Shimizu et al. 2016). The results indicated the reasonable volume for a 257-kDa complex and outlined the structure of the 1:2 complex in solution (Fig. 5, “Molecular Size” and “Outline”). The interactions within the large complex were further analyzed by X-link MSn, which detected specific interactions between KIF2 and the tubulin dimers (Fig. 5, “Interaction”). A HiRes SEC assay was also used to test the complex-forming activity of the target proteins. Interestingly, the loss of function of KIF2 mutants, both the deletion mutants (Ogawa et al. 2004) and the phosphomimetic loss of function mutants (Ogawa and Hirokawa 2015), could not form a 1:2 complex in solution (Ogawa et al. 2017).
Future direction
The combination of multiple analysis methods is now recognized as a strong approach to describe protein behavior in solution. Furthermore, the effective coupling with additional structural approaches, such as cryo-electron microscopy (cryo-EM), X-ray free-electron laser (XFEL) spectroscopy, and nuclear magnetic resonance (NMR) spectroscopy, or with the kinetic approaches such as surface plasmon resonance (SPR) and calorimetry, will lead to further advances in our understanding of protein dynamics in solution.
Acknowledgements
We thank Dr. F. Arisaka for the valuable discussion about the AUC and MALS analyses. We thank Mr. E. Tsuruta, Mr. K. Kurono, Mr. K. Watanabe, and Mr. T. Tokai of Shoko Science for their help with the SEC-RI-MALS analysis. We thank Drs. S. Sakamoto, T. Nirasawa (Bruker Daltonics), and D. Higo (Thermo Fisher Scientific) for the valuable discussions about the MS analysis. We thank all members of N. Hirokawa’s laboratory for the valuable discussions and help. We thank Ms. H. Sato, Ms. H. Fukuda, Mr. T. Akamatsu, and Mr. N. Onouchi for the technical assistance. This work was supported by Grant-in-Aid for Specially Promoted Research (MEXT KAKENHI grant numbers 18002013 and 23000013) and for Scientific Research (S)(MEXT KAKENHI grant number 16H06372) by the Ministry of Education, Culture, Sports, Science and Technology of Japan to N.H. SAXS analyses were supported by the Platform for Drug Discovery, Informatics, and Structural Life Science (PDIS) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to T.O. and N.H. (proposal number 2048). This work was partially supported by an unrestricted endowment from the Japan Electron Optics Laboratory (JEOL) and Carl Zeiss.
Author contributions
T.O. and N.H. conceived the project. T.O. performed the experiments. T.O. and N.H. discussed and wrote the paper.
Compliance with ethical standards
Conflict of interest
Tadayuki Ogawa declares that he has no conflict of interest. Nobutaka Hirokawa declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘Biomolecules to Bio-nanomachines - Fumio Arisaka 70th Birthday’ edited by Damien Hall, Junichi Takagi and Haruki Nakamura.
References
- Ando T. Directly watching biomolecules in action by high-speed atomic force microscopy. Biophys Rev. 2017;9:421–429. doi: 10.1007/s12551-017-0281-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisaka F, Kanamaru S. Protein interactions in the assembly of the tail of bacteriophage T4. Biophys Rev. 2013;5:79–84. doi: 10.1007/s12551-013-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisaka F, Yap ML, Kanamaru S, Rossmann MG. Molecular assembly and structure of the bacteriophage T4 tail. Biophys Rev. 2016;8:385–396. doi: 10.1007/s12551-016-0230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/S0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Ghirlando R, Mutskova R, Schwartz C. Enrichment and characterization of ferritin for nanomaterial applications. Nanotechnology. 2016;27:045102. doi: 10.1088/0957-4484/27/4/045102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LM, Gilbert GA (1973) [11] Sedimentation velocity measurement of protein association. Meth Enzymol 27:273–296. 10.1016/S0076-6879(73)27014-3 [DOI] [PubMed]
- Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- Igarashi N, Shimizu N, Koyama A, et al. New high-brilliance beamline BL-15A of the Photon Factory. J Phys Conf Ser. 2013;425:072016. doi: 10.1088/1742-6596/425/7/072016. [DOI] [Google Scholar]
- Ishii R, Isogaya K, Seto A, et al (2012) Structure of a dominant-negative helix-loop-helix transcriptional regulator suggests mechanisms of autoinhibition. EMBO J 31(11):2541–2552. 10.1038/emboj.2012.77 [DOI] [PMC free article] [PubMed]
- Kodera N, Yamamoto D, Ishikawa R, Ando T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010;468:72–76. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical ultracentrifugation in biochemistry and polymer science. Cambridge, UK: Royal Society of Chemistry; 1992. [Google Scholar]
- Lu C, Liu D, Liu H, Motchnik P. Characterization of monoclonal antibody size variants containing extra light chains. MAbs. 2013;5:102–113. doi: 10.4161/mabs.22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Hirokawa N. Microtubule destabilizer KIF2A undergoes distinct site-specific phosphorylation cascades that differentially affect neuronal morphogenesis. Cell Rep. 2015;12:1774–1788. doi: 10.1016/j.celrep.2015.08.018. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Nitta R, Okada Y, Hirokawa N. A common mechanism for microtubule destabilizers—M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell. 2004;116:591–602. doi: 10.1016/S0092-8674(04)00129-1. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Saijo S, Shimizu N, Jiang X, Hirokawa N. Mechanism of catalytic microtubule depolymerization via KIF2-tubulin transitional conformation. Cell Rep. 2017;20:2626–2638. doi: 10.1016/j.celrep.2017.08.067. [DOI] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. Analytical ultracentrifugation as a tool for studying protein interactions. Biophys Rev. 2013;5:159–171. doi: 10.1007/s12551-013-0106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, Yatabe K, Nagatani Y, Saijyo S, Kosuge T, Igarashi N. Software development for analysis of small-angle x-ray scattering data. AIP Conf Proc. 2016;1741:050017. doi: 10.1063/1.4952937. [DOI] [Google Scholar]
- Shipley K, Hekmat-Nejad M, Turner J, et al. Structure of a kinesin microtubule depolymerization machine. EMBO J. 2004;23:1422–1432. doi: 10.1038/sj.emboj.7600165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchihashi T, Iino R, Ando T, Noji H. High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase. Science. 2011;333:755–758. doi: 10.1126/science.1205510. [DOI] [PubMed] [Google Scholar]
- Uchiyama S, Arisaka F, Stafford WF, Laue T, editors. Analytical ultracentrifugation. Tokyo: Springer; 2016. [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/S0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Wyatt PJ. Submicrometer particle sizing by multiangle light scattering following fractionation. J Colloid Interface Sci. 1998;197:9–20. doi: 10.1006/jcis.1997.5215. [DOI] [PubMed] [Google Scholar]