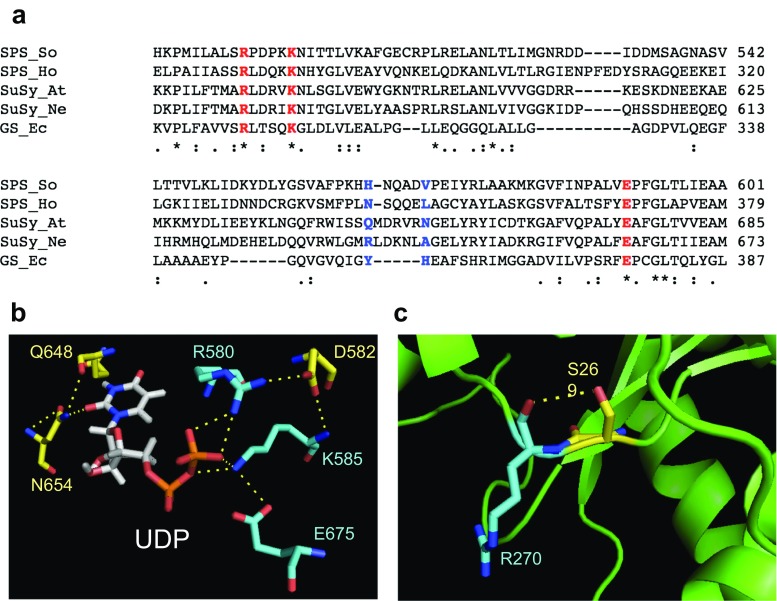
Fig. 1.
Comparison of residues in glycosyltransferases group. a Multiple sequence alignment of SPS in sugarcane (SoSPS1), SPS in non-photosynthetic bacterial H. orenii (HoSPS), sucrose synthase in A. thaliana (AtSuSy), sucrose synthase in bacteria N. europaea (NeSuSy), and glycogen synthase from E. coli (EcGS). The three highly conserved and critical residues that consist of arginine, lysine, and glutamic acid are shown in red. The residues that are predicted to be involved in nucleotide sugar interaction is shown in blue. b The UDP binding site residues at positions corresponding to Arg-580, Asp-582, Lys-585, and Glu-675 for diphosphate moiety binding and Gln-648 and Asn-654 for uridine moiety binding of A. thaliana sucrose synthase (PDB ID: 3S28). c Predicted interaction hydrogen bonding between Ser-269 and Arg-270 in bacterial SPS (PDB ID: 2R60)