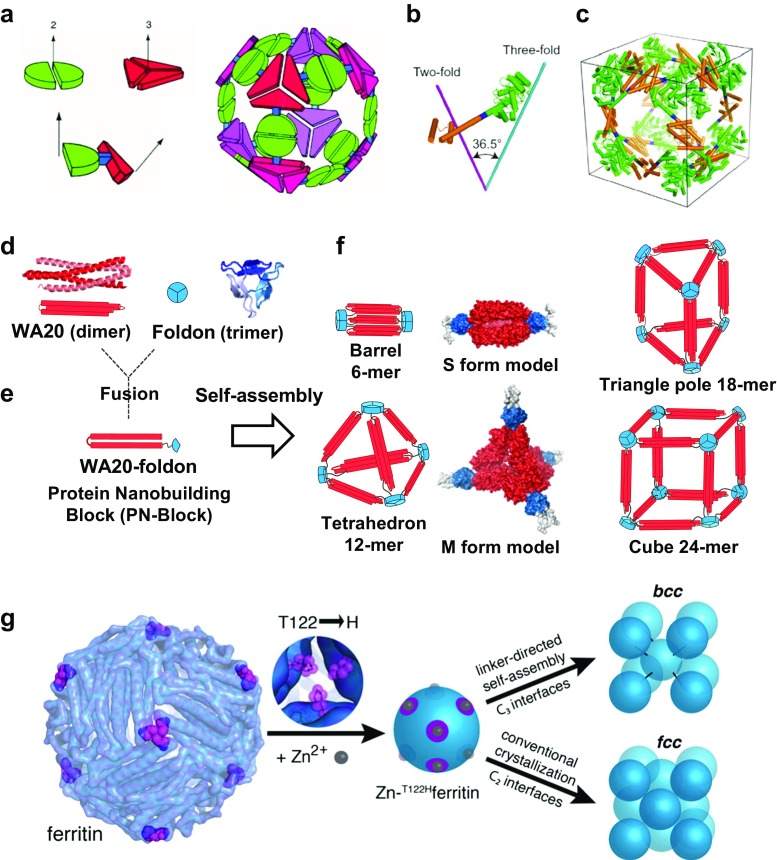
Fig. 4.
Symmetrical self-assemblies of nanoscale building blocks. a Nanohedra strategy: a general strategy for designing fusion proteins that assemble into symmetric nanostructures (Padilla et al. 2001). The green semicircle represents a natural dimeric protein (i.e., a protein that associates with one other copy of itself), whereas the red shape represents a trimeric protein. The symmetry axes of the natural oligomers are shown. The two natural proteins are combined by genetic methods into a single fusion protein. Each of the original natural proteins serves as an “oligomerization domain” in the designed fusion protein. A fusion protein is shown to illustrate that the oligomerization domains can be joined rigidly at a geometric angle. The fusion proteins self-assemble into a cubic cage of a nanohedral structure. Reprinted with permission from Padilla et al. (2001); copyright © 2001, NAS. b, c Models of the engineered fusion protein and its assembled cage structure (Lai et al. 2014). b The designed fusion protein, with trimeric KDPGal aldolase (green), the four-residue helical linker (blue) and the dimeric domain of FkpA protein (orange) shown with lines for the three-fold (cyan) and two-fold (magenta) symmetry axes. c Model of the intended 24-subunit cage with octahedral symmetry in a bounding box. Reprinted with permission from (Lai et al. 2014). Copyright © 2014, NPG. d–f Schematics of construction and assemblies of the WA20-foldon fusion protein as a protein nanobuilding block (PN-Block) (Kobayashi et al. 2015). d Ribbon representation and schematics of the intermolecularly folded dimeric WA20 (PDB ID: 3VJF) (Arai et al. 2012) shown in red, and trimeric foldon domain of T4 phage fibritin (PDB ID: 1RFO) (Guthe et al. 2004) shown in blue. e Construction of the WA20-foldon fusion protein as a PN-Block. f Schematics of designed polyhedral nanoarchitectures by expected self-assemblies of the WA20-foldon. In the stable self-assembling complexes, the WA20-foldon is expected to form highly symmetric oligomers in multiples of 6-mer because of the possible combinations of the WA20 dimer and foldon trimer. The rigid-body model structures of the S form (6-mer) and M form (12-mer) of the WA20-foldon are also shown. Reprinted with permission from Kobayashi et al. (2015); copyright © 2015, ACS. g Schematics of protein–metal–organic frameworks: metal/linker-directed self-assembly of ferritin into 3D crystals (Sontz et al. 2015). Surface-exposed binding sites for Zn2+ (gray spheres) are engineered at the C 3 pores (magenta) through the T122H mutation. In the presence of ditopic organic linkers, the resulting T122Hferritin variant is expected to form a bcc lattice. Reprinted with permission from Sontz et al. (2015); copyright © 2015, ACS