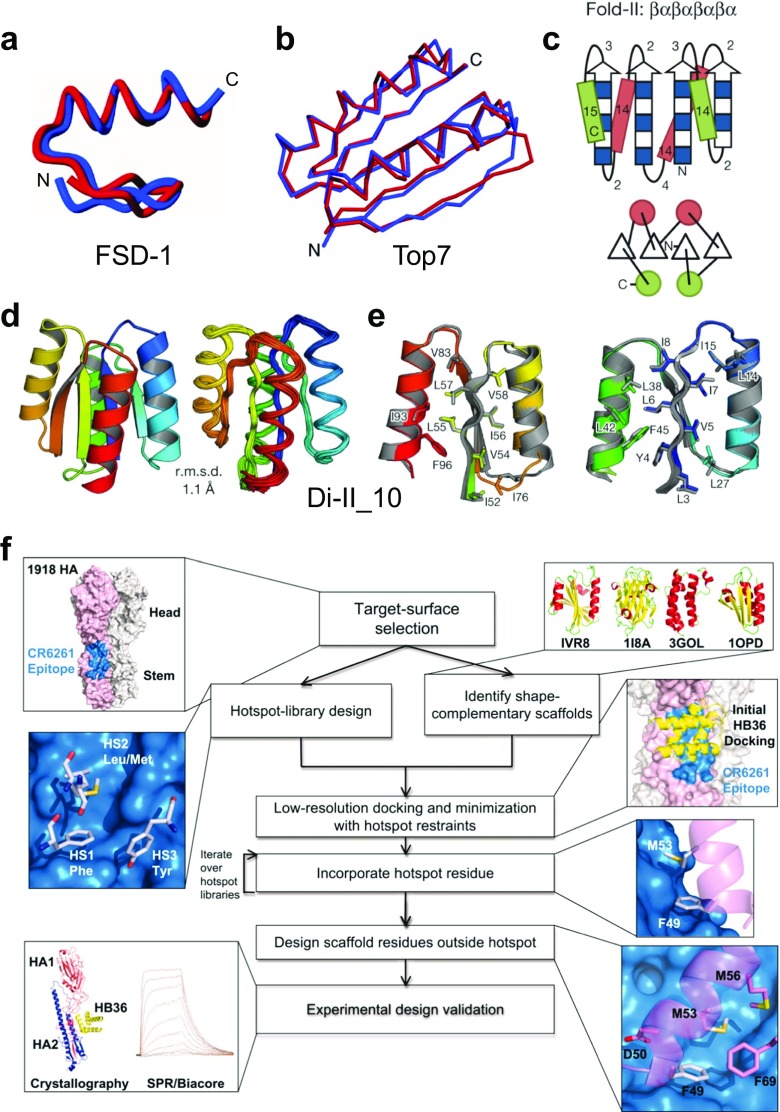
Fig. 5.
Computational design of de novo proteins. a Comparison of the FSD-1 (full sequence design 1) structure (blue) (PDB ID: 1FSD) and the design target (red) (Dahiyat and Mayo 1997). The best-fit superposition of the restrained energy minimized average NMR structure of FSD-1 and the backbone of Zif268. Reprinted with permission from Dahiyat and Mayo (1997); copyright © 1997, AAAS. b Comparison of the computationally designed model (blue) to the solved X-ray structure (red) of Top7 (PDB ID: 1QYS) (Kuhlman et al. 2003). Cα overlay of the model and structure in stereo (backbone RMSD = 1.17 Å). Reprinted with permission from Kuhlman et al. (2003); copyright © 2003, AAAS. c Secondary structure lengths from the rules for Fold-II, Rossmann2 × 2 fold. In the upper illustrations, numbers represent the loop lengths following from the rules relating local structures to tertiary motifs (Koga et al. 2012). Strand lengths are represented by filled and open boxes. The filled boxes represent pleats coming out of the page, and the open boxes represent pleats going into the page. In the lower illustrations, the design topologies are represented with circles (helices) and triangles (strands) connected by solid lines (loops). d Comparison of overall topology of Di-II_10. Design models (left) and NMR structures (right) (PDB ID: 2LV8); the Cα root mean squared deviation (r.m.s.d.) between them is indicated. e Comparison of core side-chain packing in superpositions of design models (rainbow) and NMR structures (gray). The left and right panels show close-up views of the core packing and correspond to the left and right portions of the structures shown in (d). Reprinted with permission from Koga et al. (2012); copyright © 2012, NPG. f Flow chart illustrating the key steps in the computational design of novel binding proteins. The thumbnails illustrate each step in the creation of binders that target the stem of the 1918 hemagglutinin (HA). Reprinted with permission from Fleishman et al. (2011); copyright © 2011, AAAS