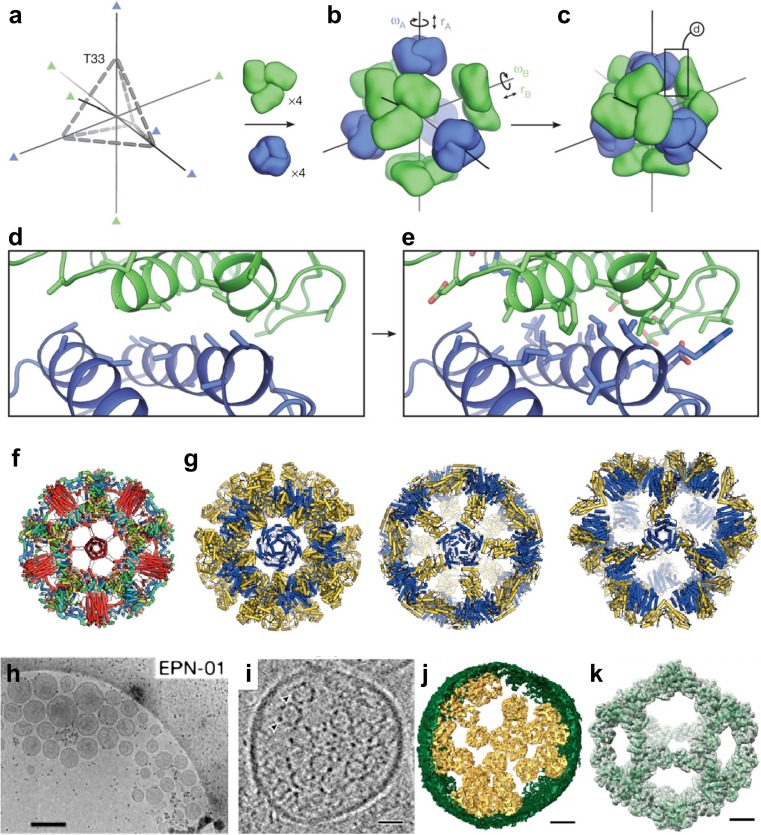
Fig. 6.
Computational design of various self-assembling protein complexes. a–e Overview of the computational design method of co-assembling multi-component protein nanomaterials (King et al. 2014). a The T33 architecture comprises four copies each of two distinct trimeric building blocks (green and blue) arranged with tetrahedral point group symmetry (24 total subunits; triangles indicate three-fold symmetry axes). b Each building block has two rigid-body degrees of freedom, one translational (r) and one rotational (ω), that are systematically explored during docking. c The docking procedure, which is independent of the amino acid sequence of the building blocks, identifies large interfaces with high densities of contacting residues formed by well-anchored regions of the protein structure. The details of such an interface, boxed, are shown in (d). e Amino acid sequences are designed at the new interface to stabilize the modeled configuration and to drive co-assembly of the two components. Reprinted with permission from King et al. (2014); copyright © 2014, NPG. f, g Designed self-assembling nanocages. f A one-component hyperstable icosahedron with a de novo helical bundle (red helices) fused in the center of the face (Hsia et al. 2016). g Two-component megadalton-scale icosahedra (Bale et al. 2016). The two components of each are colored in blue and yellow. Reprinted with permission from Huang et al. (2016); copyright © 2016, NPG. h–k Enveloped protein nanocages (EPNs) comprise cell-derived membrane envelopes containing multiple protein nanocages (Votteler et al. 2016). h Representative cryo-EM images showing extracellular vesicles/EPNs in culture supernatants from 293 T cells that expressed EPN-01. i Central slice from a cryo-EM tomographic reconstruction of a released EPN. Two internal protein nanocages are marked with arrowheads. j Isosurface model of the 3D cryo-EM reconstruction from (i). The EPN membrane is green and individual protein nanocages are gold. k Single-particle cryo-EM reconstruction of the nanocages released from EPNs following detergent treatment. Charge density from the 5.7 Å resolution electron microscopy reconstruction is shown in gray (contoured at 4.5σ). The I3–01 computational design model9 (green ribbon) was fitted into the density as a rigid body. Scale bars (h) 300 nm, (i, j) 25 nm, (k) 5 nm. Reprinted with permission from Votteler et al. (2016); copyright © 2016, NPG