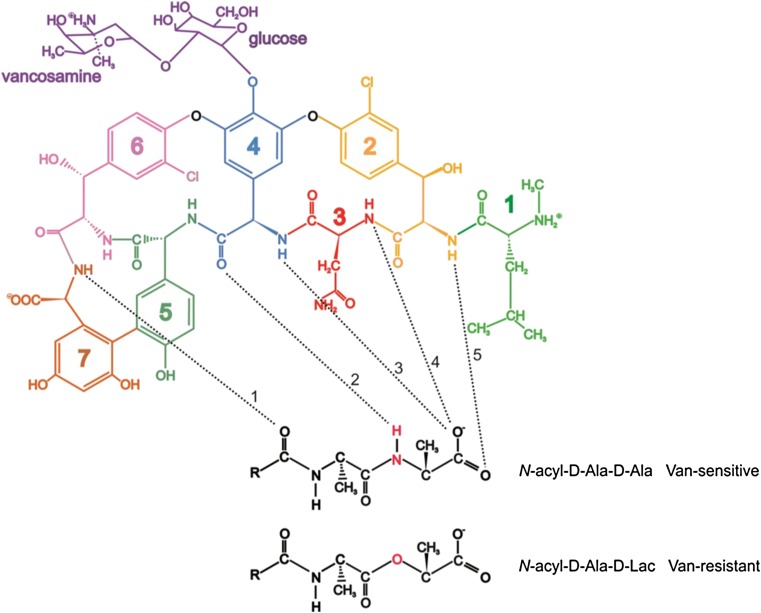
Fig. 3.
Schematic representation of vancomycin (top left) adapted and redrawn from Phillips-Jones et al. (2017a) showing the vancosamine - glucose disaccharide (purple) attached to a heptapeptide (green, N-methyl-D-leucine (residue 1); gold, m-chloro-β-hydroxy-D-tyrosine (residue 2); red, asparagine (residue 3); blue, D-phenyl glycine (residue 4); green-grey, p-hydroxy-D-phenylglycine (residue 5); pink, m-chloro- β-hydroxy-D-tyrosine (residue 6) and dark orange, m,m-dihydroxy-L-phenylglycine (residue 7)). Vancomycin binding to its sensitive target sequence (D-Ala-D-Ala) in bacterial peptidoglycan via five hydrogen bonds is shown by the dashed black lines. Hydrogen bond 2 is formed from residue 4 of vancomycin and the N-H group in D-Ala-D-Ala shown in red (middle structure). This hydrogen bond is not formed with the peptidoglycan of vancomycin-resistant bacteria that contain D-Ala-D-Lactate instead of D-Ala-D-Ala (bottom right) resulting in a total of only four hydrogen bonds for vancomycin binding which results in a 1000-fold reduced affinity of the glycopeptide for the peptidoglycan—essentially, resistance to the antibiotic