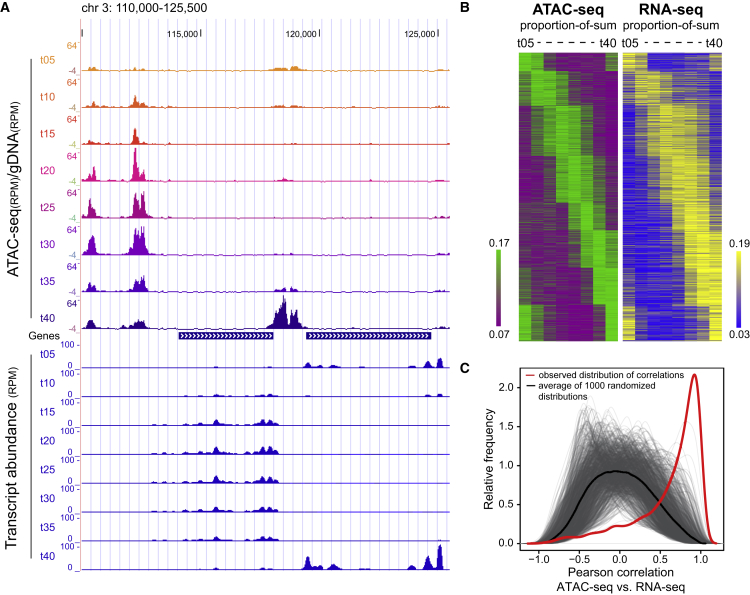
Figure 3.
Chromatin Accessibility Positively Correlates with mRNA Abundance of the Downstream Gene
(A) UCSC genome browser screenshot (chr3: 110,000–125,500) of ATAC-seq chromatin accessibility profiles (top) and corresponding RNA-seq profiles (bottom; only reads mapping to the sense strand are displayed).
(B) Heatmaps displaying relative chromatin accessibility and mRNA abundance profiles of the downstream gene through eight stages of intraerythrocytic development. Relative accessibility in ATAC-seq peaks located in divergent or tandem intergenic regions was calculated as a proportion of sum of quantile normalized RPKM values over the time points and clustered by k-means using the 1-Pearson correlation distance metric. The relative transcript abundances of the downstream located genes, expressed as proportion of sum of sense strand RPKM values over the time points, were plotted in the same order. Color scales range from the 20th to the 80th percentile for both datasets.
(C) Observed (red line) and 1,000 randomized distributions (1,000 gray lines with mean of all randomizations as black line) of Pearson correlation coefficients between relative chromatin accessibility and mRNA abundance of the downstream gene (as in B) plotted as smoothened kernel density distribution.