Abstract
In this issue of Structure, McCoy et al. (2016) describe the 2.55-Å X-ray structure of the outward-facing occluded conformation of the Bacillus cereus maltose transporter MalT. This structure represents the penultimate piece needed to complete the picture of the transport cycle of the glucose superfamily of membrane-spanning EIIC components.
The glucose-fructose-lactose (GFL) superfamily is the largest and physiologically most important superfamily of the prokaryotic phosphoenolpyruvate(PEP): sugar phosphotransferase system (PTS). The mechanism of PTS-mediated sugar uptake is unusual in that this system tightly couples sugar transport to sugar phosphorylation in a “group translocation” process (Saier et al., 2005; Västermark and Saier, 2014; Saier, 2015). In earlier reports, structures of two PTS transporters, the ChbC diacetylchitobiose (GlcNAc-β-1,4- GlcNAc, in which GlcNAc stands for “N-acetyl glucosamine”) group translocator of the GFL superfamily and the UlaA L-ascorbate group translocator of the ascorbate-galactitol (AG) superfamily had been solved (McCoy et al., 2015; Cao et al., 2011; Luo et al., 2015). The ChbC structure revealed the inward-facing occluded conformation (McCoy et al., 2015; Cao et al., 2011), whereas the UlaA structures captured the occluded and outward-facing conformations (Luo et al., 2015). UlaA, however, could not be considered directly relevant to a member of the GFL superfamily, because these two superfamilies are believed to have evolved independently of each other (Saier et al., 2005) and have very different structures.
The structure solved by McCoy et al. (2016), described in this issue of Structure, represents the outward-facing occluded conformation of the maltose (glucosyl α-1,4-glucose) transporter MalT, which belongs to the GFL superfamily. This structure is the key to understanding the complete transport cycle from outward occluded to inward occluded and confirms the model that was proposed based on the earlier ChbC structure. Overall, MalT structural work reported here and the complementary molecular dynamics (MD) simulations support the elevator car mechanism of membrane transport. In this mechanism, the transport domain of a transporter protein has two gates: one used for initial substrate binding on one side of the membrane and the other for substrate release on the other side of the membrane. Here, the substrate is completely occluded while within the membrane, and transport happens via a major movement of the entire transporter domain across the lipid bilayer. This elevator mechanism has been somewhat controversial; the MalT structure in the outward occluded conformation reported here will serve to remove some of the contention in the field given that it captures MalT in a state which is in full agreement with the model.
Now that we have three out of four structures relevant to the elevator car mechanism, we want to take a moment to examine several aspects of these structures and their functional implications in more detail. The first in-depth topic we want to consider is the proposed conformational strain of the periplasmic β-structure in ChbC. The ChbC transporter has two antiparallel periplasmic β strands. We believe that the structurally determined form of ChbC is captured in an inward- facing occluded state, although it has been proposed that the β strands in ChbC are not in their native conformation (McCoy et al., 2015; Cao et al., 2011). We find that the periplasmic β-structure of ChbC displays hydrogen bonding angles and distances within the normal range. In MalT, rigid body rotation translocates the sugar substrate by 20 Å, resulting in the extended transmembrane segment (TMS) 3–4 interconnector, which forms an additional antiparalell β-structure. In the UlaA structure, conformational energy is not affected by the hydrogen bonds of the mini β-clusters between the open and the outward state, as can be shown by juxtapositioning the periplasmic clusters of the two structures. Regardless of whether this is a transporter- or state-specific difference, we argue that the periplasmic β-structure of ChbC’s dimerization domain displays angles typically found in secondary structures and seems not to be under conformational strain.
The second issue that we would like to comment on is whether the substrate binding site in these transporters is solvent exposed and what the role of this might be, given that the elevator car mechanism of membrane transport assumes that the substrate is fully occluded from the solvent. Diacetylchitobiose viewed from the cytoplasmic face of one monomer of ChbC in the inward-facing occluded state shows that a part of the reducing GlcNAc moiety is exposed and can be seen through a small opening in the surface lining encircled by Met33, Pro177, and Thr253 (McCoy et al., 2015; Cao et al., 2011). Outward-facing occluded MalT’s periplasmic cavity opening appears too narrow for substrate access, but inwardly occluded ChbC’s trans-protomer loop only partially shields the substrate from cytoplasmic solvent (McCoy et al., 2015; Cao et al., 2011). The existence of this solvent-accessible opening can be used to explain how the phosphate group is transferred from the ChbB component to the ChbC substrate. ChbC phosphorylates diacetylchitobiose on the C6′-hydroxyl group of the non-reducing moiety, which must be close to Glu334 and His250. However, we now see that MalT facilitates maltose diffusion in the absence of the IIB protein (McCoy et al., 2016), and the small cavity opening in MalT might play different roles.
By comparing outward-facing open and occluded UlaA using Δ-distance maps (Nishikawa and Ooi, 1974), it is clear that TMS7 moved compared to the rest of the structure. TMS7 is part of the V motif, the feature that is shared between the structures. Regarding conformational stress, the fact that two TMSs are pulled up from the interior side of the membrane, making them reentrant-like, the increased amount of periplasmic β-structure, and the Ramachandran statistics for the new structure indicate that the outward-facing occluded structure is in a high energy state. This could drive the inward transport process, especially in the absence of the other PTS proteins. The rigid body model was first proposed based on a re-entrant hairpin similar to the one found in a secondary carrier, GltPh (Yernool et al., 2004), but re-entrant hairpins are common features of transporters, and in retrospect, even though the rotating mechanism proposed in the ChbC paper (Cao et al., 2011) now appears confirmed, GltPh was too distant a candidate from which to import mechanistic predictions. Furthermore, how is the substrate released if it is held back from the periplasm by a loop from the dimerization protomer that might not be rotatable in a way analogous to TMS7? It was proposed by Cao et al. (2011) that the substrate was not solvent accessible from the cytoplasm, but careful examination of the structure shows that it could be, perhaps suggesting that substrate phosphorylation can occur in the inward occluded state. Because these proteins transport only weakly in the absence of the other PTS proteins and have larger substrate cavities, it seems likely that ChbC can bind some form of trisaccharide, but the phosphorylation process might be less efficient. The top ChbC docking mode of the trisaccharide displays an affinity of −9.6 kcal/mol, preserving the orientation of the phosphorylation site. How this observation translates to MalT remains unclear, but it might be easier for maltotriose to bind to the outward form of the transporter than it is for it to get phosphorylated and released on the inside.
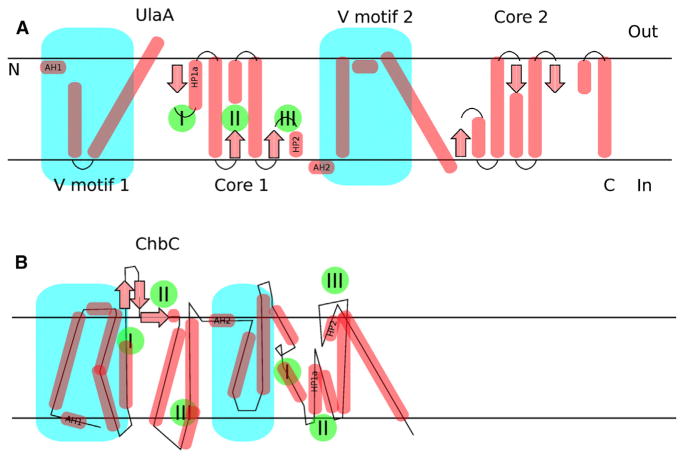
In closing, the membrane-spanning EIIC components of the PTS contain secondary structural elements that can be mapped between UlaA and ChbC (Figure 1). This suggests that the spatial configurations displayed by the known conformational states of these proteins can be achieved by rearranging the cores and V motifs (the “skull and crossbones”) in both proteins. Δ-distance maps (Nishikawa and Ooi, 1974) to compare outward occluded (P21B) and outward open (C2A) states mainly confirmed the original model (Luo et al., 2015), but also revealed some important differences (data not shown; available upon request). Consequently, the TMS7 rotational mechanism of periplasmic mouth cavity shrinking proposed from MalT simulations might be validated using crystallographic data from UlaA. One of the next challenges in this field will be to extend the approach of complementing X-ray crystallography with MD to assess the inward occluded to inward open mechanism. We enjoyed McCoy et al. (2016) and hope you do too!
Figure 1. Mapping between Secondary Structural Elements of UlaA and ChbC or MalT.
(A) The V motifs of UlaA (turquoise) can be mapped onto ChbC or MalT, both having AH1 and 2 at their N-termini and before the V motif domain. However, whereas UlaA shows a repeat sequence (two homologous domains; Saier et al., 2005), obvious from the 3D structure, ChbC does not, and the two proteins do not exhibit the same 3D structural fold. Each core motif of UlaA has three hairpins/broken helices (marked with green circles/roman numerals I, II, and III) at the start, middle, and end of each core unit. It is currently accepted that the C-terminal end is located in the cytoplasm, based on the “positive inside rule” (von Heijne, 1989), and in cases in which cytosolic UlaB homologs are physically linked to the C-termini of UlaA homologs. (B) Corresponding elements in different positions can be found in ChbC or MalT. The fact that the V and core motif nomenclature can be applied to ChbC or MalT suggests secondary structural similarity, although this is not apparent on the primary or tertiary sequence/structure levels. AH, amphipathic helix; HP, hairpin.
Acknowledgments
This work was supported by NIH Grant GM077402 (to M.H.S.).
References
- Cao Y, Jin X, Levin EJ, Huang H, Zong Y, Quick M, Weng J, Pan Y, Love J, Punta M, et al. Nature. 2011;473:50–54. doi: 10.1038/nature09939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P, Yu X, Wang W, Fan S, Li X, Wang J. Nat Struct Mol Biol. 2015;22:238–241. doi: 10.1038/nsmb.2975. [DOI] [PubMed] [Google Scholar]
- McCoy JG, Levin EJ, Zhou M. Biochim Biophys Acta. 2015;1850:577–585. doi: 10.1016/j.bbagen.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy JG, Ren Z, Stanevich V, Lee J, Mitra S, Levin EJ, Poget S, Quick M, Im W, Zhou M. Structure. 2016;24(this issue):956–964. doi: 10.1016/j.str.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K, Ooi T. J Theor Biol. 1974;43:351–374. doi: 10.1016/s0022-5193(74)80066-4. [DOI] [PubMed] [Google Scholar]
- Saier MH., Jr J Mol Microbiol Biotechnol. 2015;25:73–78. doi: 10.1159/000381215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Hvorup RN, Barabote RD. Biochem Soc Trans. 2005;33:220–224. doi: 10.1042/BST0330220. [DOI] [PubMed] [Google Scholar]
- Västermark A, Saier MH., Jr Curr Opin Microbiol. 2014;18:8–15. doi: 10.1016/j.mib.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- Yernool D, Boudker O, Jin Y, Gouaux E. Nature. 2004;431:811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]