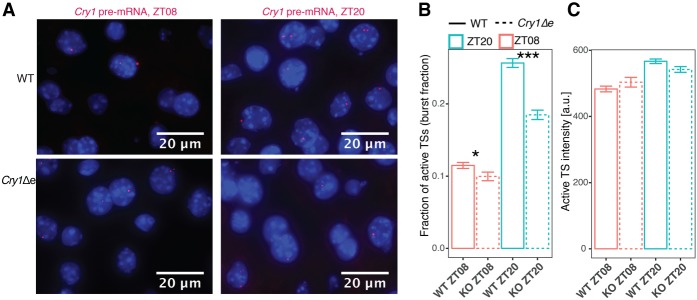
Figure 6.
The oscillatory Cry1 promoter–enhancer loop modulates Cry1 transcriptional bursting. (A) smRNA-FISH against Cry1 pre-mRNA in the livers of wild-type (top) and Cry1Δe (bottom) animals at ZT08 (left) and ZT20 (right). Burst fractions (B) and burst intensities (C) measured from images of smRNA-FISH performed against Cry1 pre-mRNA in Cry1Δe (dashed) and wild-type (solid) livers at ZT08 (red) and ZT20 (blue). Burst fraction is the number of active transcription sites in each nucleus divided by the ploidy. (B,C) Shown are the means and standard errors over nuclei collected and pooled from two animals in each of the four conditions (individual animals are analyzed in Supplemental Fig. S9C,D). n = 2191 wild-type ZT08 nuclei; n = 983 Cry1Δe ZT08 nuclei; n = 2150 wild-type ZT20 nuclei; n = 1473 Cry1Δe ZT20 nuclei. In B, (*) P < 0.05; (***) P < 0.001, t-test. In C, differences between genotypes are not significant.