Figure 6.
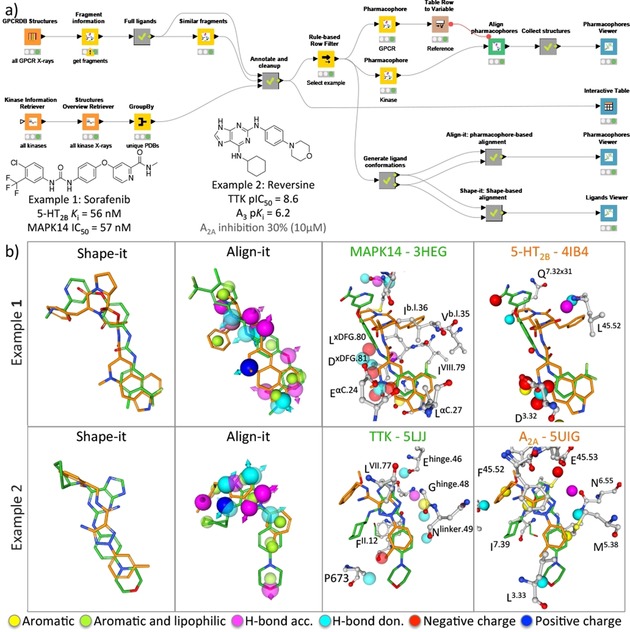
A structure‐based ligand repurposing workflow (A) that searches for KripoDB26 pharmacophore similarities between GPCRs and kinases. Two examples (B) of binding site similarities between the 5‐HT2B receptor and MAPK14 kinase, and the adenosine A2A receptor and the TTK kinase are presented and described in the main text. The aligned kinase and GPCR structures based on the alignment of the KRIPO pharmacophores are shown in 3D using the Proteins and Ligands viewer (for clarity purposes the lipophilic pharmacophore features are hidden). Only residues within 3.5 Å of the ligands are depicted and labeled according to the Ballesteros–Weinstein45 and KLIFS24 numbering scheme for GPCRs and kinases, respectively. Complementary shape‐based and pharmacophore‐based assessment of the ligands using the KNIME‐enabled Silicos‐it25 tools Shape‐it and Align‐it are performed and compared in the Ligands viewer and Pharmacophore viewer, respectively.
