Figure 1.
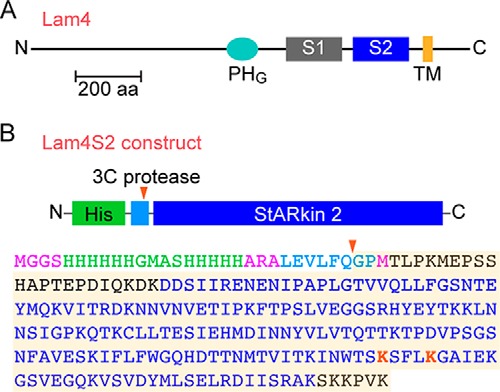
Lam4 and the Lam4S2 construct. A, schematic illustration of Lam4 (Yhr080c) indicating the PH-like GRAM domain (PHG), the two StARkin domains (S1 and S2), and transmembrane (TM) domain. The latter is strongly predicted by the OCTOPUS membrane protein topology prediction web server (http://octopus.cbr.su.se/ (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.)) (48). The protein is 1345 amino acids long (∼150 kDa) and represented roughly to scale according to the indicated 200-amino acid (aa) scale bar. The transmembrane domain anchors Lam4 to the ER membrane such that its N terminus is in the cytoplasm. B, sequence of the Lam4S2 construct used in our studies. The second StARkin domain was expressed as a His-tagged protein in E. coli. The His tag (green) was attached via a linker sequence (light blue) containing a 3C protease cleavage site. The red arrowhead indicates the cleavage site. The amino acid sequence of the construct is color-coded to indicate the His tag (green), consensus sequence for 3C protease cleavage (light blue), the StARkin 2 domain (navy blue; as defined in Ref.17), and lysine residues (red) that were targeted by site-directed mutagenesis in our studies. Residues in black are part of the native Lam4 sequence, extending on either side of the StARkin 2 domain; residues in pink are part of the design of the construct. The shaded sequence corresponds to the Lam4S2 protein (predicted molecular mass, 22.7 kDa) used for crystallography with residue numbering starting with the first glycine. In this numbering scheme, the highlighted lysine residues are Lys-163 and Lys-167. The fourth residue in the shaded sequence, Thr-4, corresponds to Thr-946 in native Lam4.
