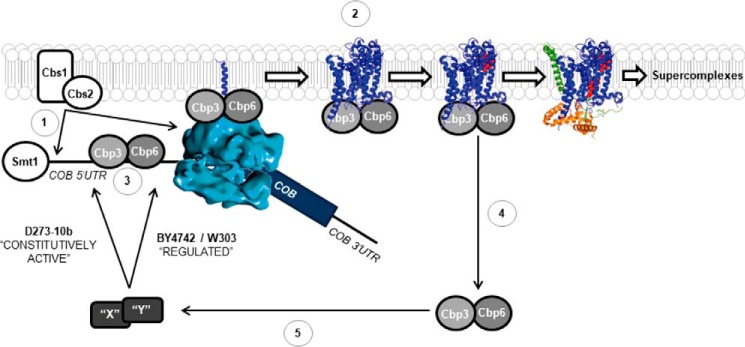
Figure 8.
Model showing a mechanism for the differential regulation of Cytb synthesis by Cbp3 and Cbp6 in distinct laboratory yeast strains. Translational activation of the COB mRNA depends on Cbs1 and Cbs2 (8, 9) (1). Cbp3 and Cbp6 interact with the ribosomal tunnel exit (14) and with newly synthesized Cytb to promote hemylation/assembly (hemes in red) (7, 15) (2). Cbp3 interacts with the COB 5′-UTR mRNA (this work), probably to promote efficient translation (3). These interactions are present in both D273-10b and BY4742 lab strains. Cytb assembly proceeds to form the first intermediary, containing Qcr7 (orange) and Qcr8 (green) subunits, and then Cbp3/6 release from the Cytb complex (7). We propose the existence of additional factors X and Y, which in turn become active in BY4742 and W303 lab strains, allowing an efficient COB mRNA translation (5). Absence of Cbp3/6 prevents factors X and Y from becoming active, and thereby Cytb synthesis is reduced. In contrast, in D273-10b strain factors X and Y are constitutively active, even in the absence of Cbp3 and Cbp6. In addition, Smt1 physically interacts with the COB mRNA to carry over an unknown function that does not seem to be related to Cbp3/Cbp6 (20). Tertiary and quaternary structures were taken from the Protein Data Bank (code 1EZV).