Figure 2.
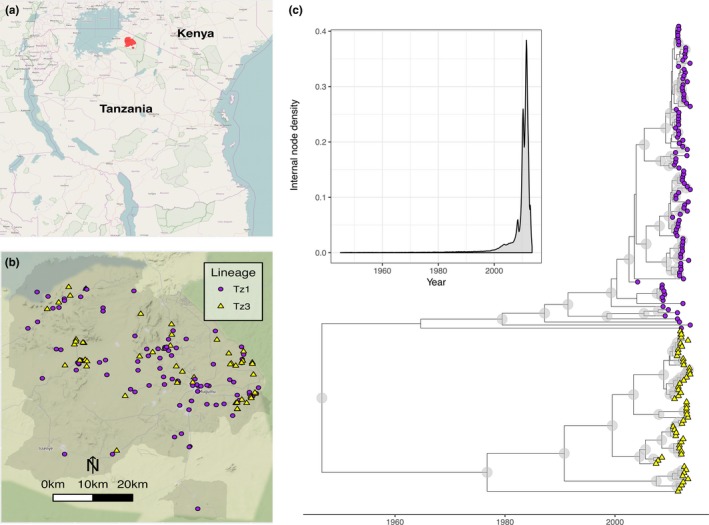
The spatial location and phylogenetic structure of 152 sequenced rabies viruses sampled from 2004 to 2013 within the Serengeti district, Tanzania. (a) The Serengeti district (red polygon) within Tanzania; (b) locations of sequenced rabies cases within the Serengeti district (grey polygon) with underlying topography (map tiles by Stamen Design, under CC BY 3.0. Data by OpenStreetMap, under ODbL.) and administrative boundaries from http://www.nbs.go.tz; (c) timescaled maximum clade credibility tree from a Bayesian phylogenetic reconstruction of whole‐genome sequences, with node posterior support >0.9 indicated by blue circles. The inset shows node density through time for the posterior set of trees, with >90% nodes occurring in the last 10 years. Maps drawn using R packages OpenStreetMap (Fellows & Stotz, 2016) ggmap (Kahle & Wickham, 2013) and maptools (Lewin‐Koh et al., 2012)
