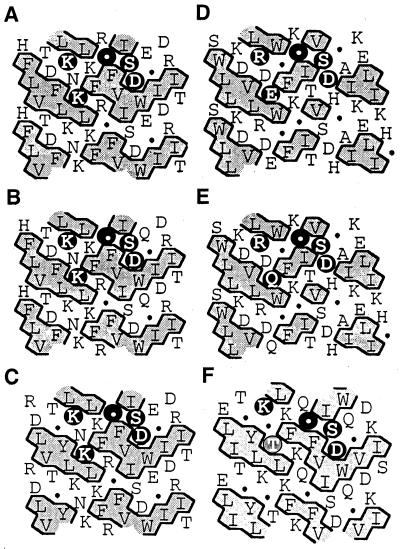
Figure 4.
Comparison of HCA of the potential domain containing the aspartyl nucleophile of maize and barley ExGases with those of four family 3 glycosidases with homology to the maize ExGase open reading frame. In the HCA plots, the sequences of proteins are arranged in a duplicated α-helical net, and clusters of contiguous hydrophobic amino acids are shaded. The amino acids conforming to the [KR]-x-[EQK]-xxxx-G-xxx-[ST]-D motif are highlighted with black circles in the upper α-helical net. The Thr residue in the Arabidopsis analysis that does not conform to the family 3 motif is shaded. Gly is represented by a black circle in these representations. A, Maize ExGase; B, barley ExGase (Hrmova et al., 1996); C, nasturtium ExGase (Crombie et al., 1998); D, Salmonella typhimurium β-glucosidase (T-cell inhibitor; D86507); E, Escherichia coli periplasmic β-glucosidase (U15049); and F, Arabidopsis ExGase (AB008271, clone MUK11).