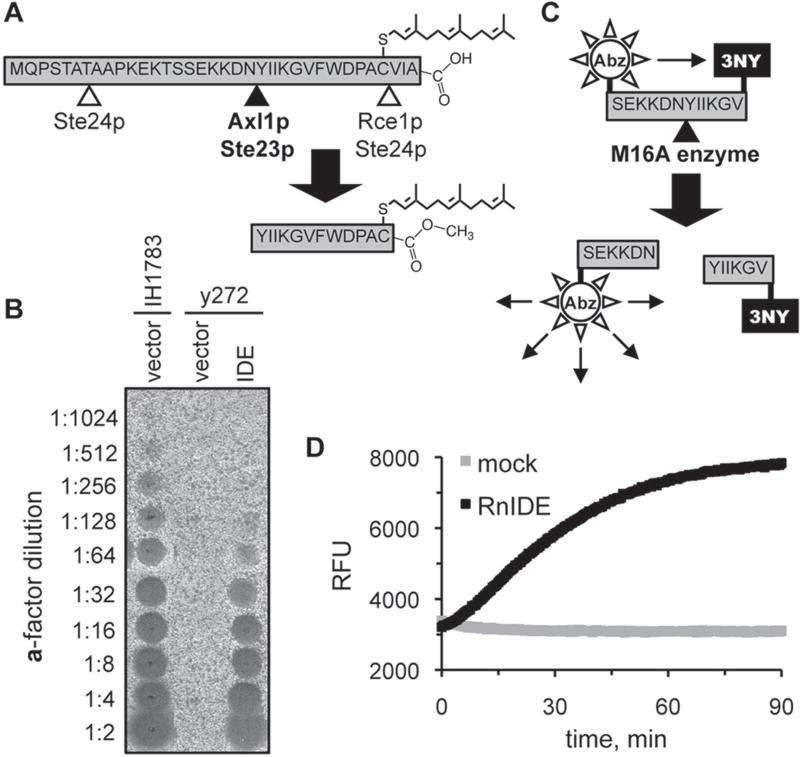
Figure 1.
Reporters of M16A enzyme activity. (A) Production of the yeast a-factor mating pheromone is dependent on the action of several proteases, including the M16A enzymes Axl1p and Ste23p. (B) Rat insulin-degrading enzyme (IDE) can substitute for the yeast M16A enzymes in a-factor production in vivo. Yeast strain y272 (MATa axl1 Δ ste23Δ) was transformed with an IDE-encoding plasmid (pWS491) or an empty vector (pRS316), and resultant strains were evaluated for their ability to produce a-factor. A wild-type MATa strain (IH1783) transformed with an empty vector (pRS316) was evaluated in parallel. The appearance of a clear spot (i.e., zone of reduced growth) within the MATα lawn indicates the presence of a-factor. (C) An internally quenched fluorogenic dodecapeptide modeled on the M16A cleavage site in the yeast a-factor precursor. The NH2-terminal fluorophore is aminobenzoic acid (Abz), and the COOH-terminal quenching group is 3-nitro-tyrosine (3NY). (D) Progress curves demonstrating time-dependent fluorescent output in the presence or absence of recombinant rat IDE. The reactions contained rat IDE (10 µg/ml; 87.7 nM) or enzyme storage buffer (mock). RFU, relative fluorescence units.