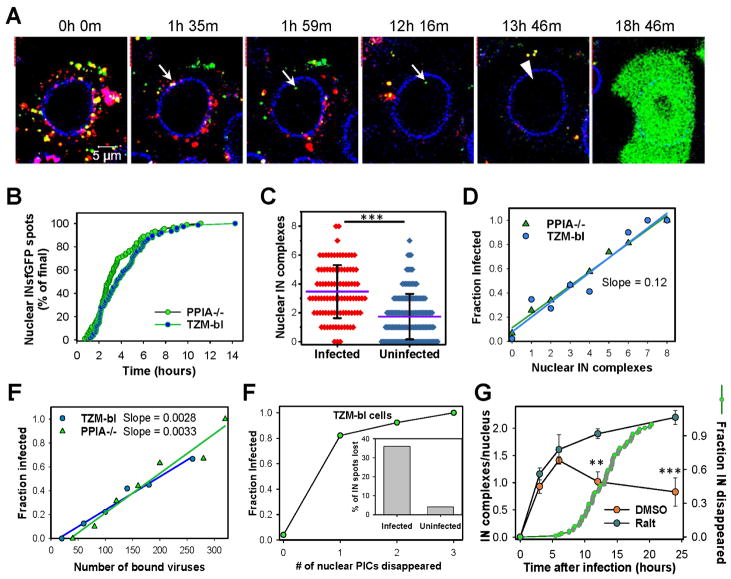
Fig. 2. Nuclear INsfGFP complexes are likely to establish infection.
(A) EBFP2-Lamin expressing TZM-bl cells infected with INsfGFP/CypA-DsRed labeled pseudoviruses encoding for eGFP were continuously imaged, starting at 0 h.p.i. An arrow marks a single particle docking at the NE, losing CypA-DsRed and entering the nucleus. An arrowhead shows subsequent disappearance of the nuclear INsfGFP signal prior to eGFP reporter expression. Scale bar 5 μm. (B) Kinetics of live-cell nuclear import of INsfGFP complexes in TZM-bl and PPIA−/− cells infected with INsfGFP/CypA-DsRed labeled viruses (≥5 independent experiments). (C and D). Analysis of the number of nuclear IN complexes in infected (eGFP+) vs. uninfected (eGFP−) cells and correlation between the nuclear complexes and the probability of eGFP expression after inoculation with pseudoviruses at MOI 0.2. (E) Correlation between the number of INsfGFP puncta associated with each cell after incubation for 10 min at 37°C and infection. (F) The nuclear INsfGFP complex disappearance events were detected and correlated with the infection outcome. Inset: A fraction of IN-spots that disappeared in infected vs. uninfected cells. Total cells analyzed from ≥3 independent experiments in panels C–F: GFP+ (PPIA−/−, n=97; TZM-bl, n=74) and GFP- (PPIA−/−, n=154; TZM-bl, n=130). (G) Time course of nuclear detection of INsfGFP spots in TZM-bl cells after infection with pR9ΔE/VSV-G pseudovirus labeled with INsfGFP and CypA-DsRed in the presence of a fully inhibitory concentration of Raltegravir (10 μM) or DMSO (control). Infection was carried out in the presence of aphidicolin (5 μM). Cells were fixed at indicated time points and the number of nuclear IN complexes measured. Error bars are SEM from 4 fields of view, from a representative of 2 independent experiments. An additional plot (right axis, green circles) shows the kinetics of nuclear INsfGFP puncta disappearance measured in live TZM-bl cells in parallel experiments (n=97; cumulative of 2 independent experiments). See related Figure S4 and movie S1.