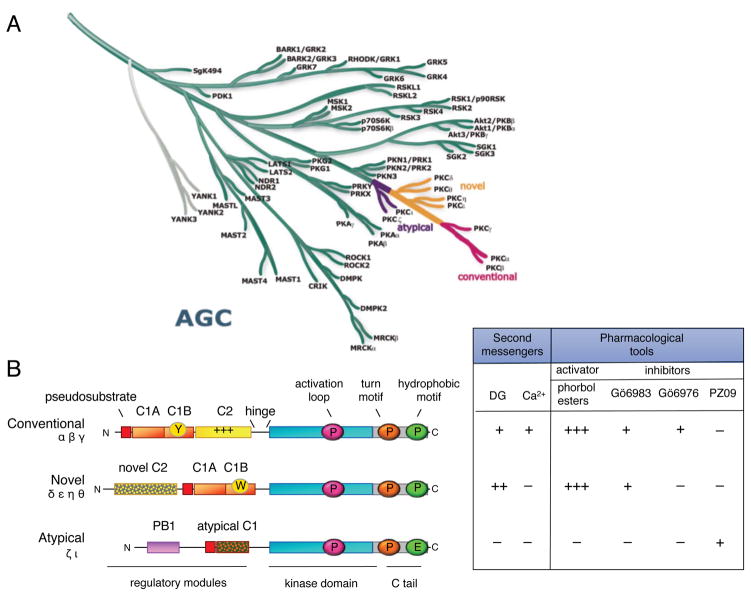
Figure 2. PKC isozymes are AGC kinases with N-terminal modules that control activity.
A. The AGC branch of the human kinome (reproduced from www.cellsignal.com/reference/kinase and courtesy of Gerard Manning) showing the position of the PKC isozymes. This branch includes Akt, p70S6 kinase and PDK-1. Most closely related to the PKC isozymes are the PKN family members that diverge first from the branch, followed by the atypical PKC isozymes (purple), the novel PKC isozymes (orange), and finally, the conventional PKC isozymes (pink), which are at the tip of the branch. B. Domain composition of PKC family members showing pseudosubstrate (red rectangle), C1 domain (orange rectangle; Y/W switch that dictates affinity for diacylglycerol-containing membranes indicated by circle in C1B domain), C2 domain (yellow rectangle; basic patch that drives binding to PIP2 indicated by ++ on domain), connecting hinge segment, kinase domain (cyan), and carboxyl-terminal tail (CT, grey rectangle). Also shown are the three priming phosphorylations: the activation loop in the kinase domain (magenta circle) and the turn motif (orange circle) and hydrophobic motif (green circle) in the carboxyl-terminal tail (note atypical PKC isozymes have Glu at phospho-acceptor position of hydrophobic motif). Table shows dependence of protein kinase C family members on second messengers (diacyglycerol (DG) and Ca2+) and pharmacological tools to activate (phorbol esters) or inhibit (Gö 6983, Gö 6976, and PZ09) PKC; +, ++, and +++ indicate relative affinity for C1 domain ligands.